Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA
- PMID: 24310612
- PMCID: PMC4049335
- DOI: 10.1126/science.1241328
Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA
Abstract
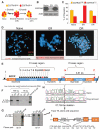
Intratumoral heterogeneity contributes to cancer drug resistance, but the underlying mechanisms are not understood. Single-cell analyses of patient-derived models and clinical samples from glioblastoma patients treated with epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) demonstrate that tumor cells reversibly up-regulate or suppress mutant EGFR expression, conferring distinct cellular phenotypes to reach an optimal equilibrium for growth. Resistance to EGFR TKIs is shown to occur by elimination of mutant EGFR from extrachromosomal DNA. After drug withdrawal, reemergence of clonal EGFR mutations on extrachromosomal DNA follows. These results indicate a highly specific, dynamic, and adaptive route by which cancers can evade therapies that target oncogenes maintained on extrachromosomal DNA.
Figures



References
-
- Garraway LA, Jänne PA. Cancer Discov. 2012;2:214–226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

