Snf2 family gene distribution in higher plant genomes reveals DRD1 expansion and diversification in the tomato genome
- PMID: 24312269
- PMCID: PMC3842944
- DOI: 10.1371/journal.pone.0081147
Snf2 family gene distribution in higher plant genomes reveals DRD1 expansion and diversification in the tomato genome
Abstract
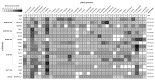
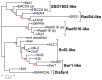
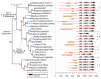
As part of large protein complexes, Snf2 family ATPases are responsible for energy supply during chromatin remodeling, but the precise mechanism of action of many of these proteins is largely unknown. They influence many processes in plants, such as the response to environmental stress. This analysis is the first comprehensive study of Snf2 family ATPases in plants. We here present a comparative analysis of 1159 candidate plant Snf2 genes in 33 complete and annotated plant genomes, including two green algae. The number of Snf2 ATPases shows considerable variation across plant genomes (17-63 genes). The DRD1, Rad5/16 and Snf2 subfamily members occur most often. Detailed analysis of the plant-specific DRD1 subfamily in related plant genomes shows the occurrence of a complex series of evolutionary events. Notably tomato carries unexpected gene expansions of DRD1 gene members. Most of these genes are expressed in tomato, although at low levels and with distinct tissue or organ specificity. In contrast, the Snf2 subfamily genes tend to be expressed constitutively in tomato. The results underpin and extend the Snf2 subfamily classification, which could help to determine the various functional roles of Snf2 ATPases and to target environmental stress tolerance and yield in future breeding.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

