The venom gland transcriptome of Latrodectus tredecimguttatus revealed by deep sequencing and cDNA library analysis
- PMID: 24312294
- PMCID: PMC3842942
- DOI: 10.1371/journal.pone.0081357
The venom gland transcriptome of Latrodectus tredecimguttatus revealed by deep sequencing and cDNA library analysis
Abstract
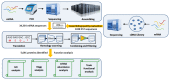
Latrodectus tredecimguttatus, commonly known as black widow spider, is well known for its dangerous bite. Although its venom has been characterized extensively, some fundamental questions about its molecular composition remain unanswered. The limited transcriptome and genome data available prevent further understanding of spider venom at the molecular level. In the present study, we combined next-generation sequencing and conventional DNA sequencing to construct a venom gland transcriptome of the spider L. tredecimguttatus, which resulted in the identification of 9,666 and 480 high-confidence proteins among 34,334 de novo sequences and 1,024 cDNA sequences, respectively, by assembly, translation, filtering, quantification and annotation. Extensive functional analyses of these proteins indicated that mRNAs involved in RNA transport and spliceosome, protein translation, processing and transport were highly enriched in the venom gland, which is consistent with the specific function of venom glands, namely the production of toxins. Furthermore, we identified 146 toxin-like proteins forming 12 families, including 6 new families in this spider in which α-LTX-Lt1a family2 is firstly identified as a subfamily of α-LTX-Lt1a family. The toxins were classified according to their bioactivities into five categories that functioned in a coordinate way. Few ion channels were expressed in venom gland cells, suggesting a possible mechanism of protection from the attack of their own toxins. The present study provides a gland transcriptome profile and extends our understanding of the toxinome of spiders and coordination mechanism for toxin production in protein expression quantity.
Conflict of interest statement
Figures






References
-
- Sebastin PA, KPe, editors (2009) Spiders of India.
-
- Kirkness EF, Haas BJ, Sun W, Braig HR, Perotti MA et al. (2010) Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc Natl Acad Sci U S A 107: 12168-12173. doi:10.1073/pnas.1003379107. PubMed: 20566863. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

