TAL effector specificity for base 0 of the DNA target is altered in a complex, effector- and assay-dependent manner by substitutions for the tryptophan in cryptic repeat -1
- PMID: 24312634
- PMCID: PMC3849474
- DOI: 10.1371/journal.pone.0082120
TAL effector specificity for base 0 of the DNA target is altered in a complex, effector- and assay-dependent manner by substitutions for the tryptophan in cryptic repeat -1
Abstract
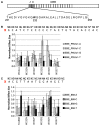
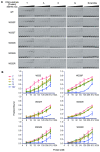
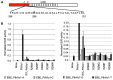
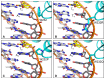
TAL effectors are re-targetable transcription factors used for tailored gene regulation and, as TAL effector-nuclease fusions (TALENs), for genome engineering. Their hallmark feature is a customizable central string of polymorphic amino acid repeats that interact one-to-one with individual DNA bases to specify the target. Sequences targeted by TAL effector repeats in nature are nearly all directly preceded by a thymine (T) that is required for maximal activity, and target sites for custom TAL effector constructs have typically been selected with this constraint. Multiple crystal structures suggest that this requirement for T at base 0 is encoded by a tryptophan residue (W232) in a cryptic repeat N-terminal to the central repeats that exhibits energetically favorable van der Waals contacts with the T. We generated variants based on TAL effector PthXo1 with all single amino acid substitutions for W232. In a transcriptional activation assay, many substitutions altered or relaxed the specificity for T and a few were as active as wild type. Some showed higher activity. However, when replicated in a different TAL effector, the effects of the substitutions differed. Further, the effects differed when tested in the context of a TALEN in a DNA cleavage assay, and in a TAL effector-DNA binding assay. Substitution of the N-terminal region of the PthXo1 construct with that of one of the TAL effector-like proteins of Ralstonia solanacearum, which have arginine in place of the tryptophan, resulted in specificity for guanine as the 5' base but low activity, and several substitutions for the arginine, including tryptophan, destroyed activity altogether. Thus, the effects on specificity and activity generated by substitutions at the W232 (or equivalent) position are complex and context dependent. Generating TAL effector scaffolds with high activity that robustly accommodate sites without a T at position 0 may require larger scale re-engineering.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

