Alcohol metabolism and epigenetics changes
- PMID: 24313160
- PMCID: PMC3860421
Alcohol metabolism and epigenetics changes
Abstract
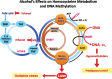
Metabolites, including those generated during ethanol metabolism, can impact disease states by binding to transcription factors and/or modifying chromatin structure, thereby altering gene expression patterns. For example, the activities of enzymes involved in epigenetic modifications such as DNA and histone methylation and histone acetylation, are influenced by the levels of metabolites such as nicotinamide adenine dinucleotide (NAD), adenosine triphosphate (ATP), and S-adenosylmethionine (SAM). Chronic alcohol consumption leads to significant reductions in SAM levels, thereby contributing to DNA hypomethylation. Similarly, ethanol metabolism alters the ratio of NAD+ to reduced NAD (NADH) and promotes the formation of reactive oxygen species and acetate, all of which impact epigenetic regulatory mechanisms. In addition to altered carbohydrate metabolism, induction of cell death, and changes in mitochondrial permeability transition, these metabolism-related changes can lead to modulation of epigenetic regulation of gene expression. Understanding the nature of these epigenetic changes will help researchers design novel medications to treat or at least ameliorate alcohol-induced organ damage.
Figures




References
-
- Alano CC, Ying W, Swanson RA. Poly(ADP-ribose) polymerase-1-mediated cell death in astrocytes requires NAD+ depletion and mitochondrial permeability transition. Journal of Biological Chemistry. 2004;279:18895–18902. - PubMed
-
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 447:407–412. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

