Inversion of a part of the numerator relationship matrix using pedigree information
- PMID: 24313900
- PMCID: PMC3878974
- DOI: 10.1186/1297-9686-45-45
Inversion of a part of the numerator relationship matrix using pedigree information
Abstract
Background: In recent theoretical developments, the information available (e.g. genotypes) divides the original population into two groups: animals with this information (selected animals) and animals without this information (excluded animals). These developments require inversion of the part of the pedigree-based numerator relationship matrix that describes the genetic covariance between selected animals (A22). Our main objective was to propose and evaluate methodology that takes advantage of any potential sparsity in the inverse of A22 in order to reduce the computing time required for its inversion. This potential sparsity is brought out by searching the pedigree for dependencies between the selected animals. Jointly, we expected distant ancestors to provide relationship ties that increase the density of matrix A22 but that their effect on A22-1 might be minor. This hypothesis was also tested.
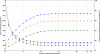
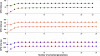
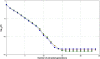
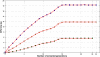
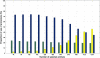
Methods: The inverse of A22 can be computed from the inverse of the triangular factor (T-1) obtained by Cholesky root-free decomposition of A22. We propose an algorithm that sets up the sparsity pattern of T-1 using pedigree information. This algorithm provides positions of the elements of T-1 worth to be computed (i.e. different from zero). A recursive computation of A22-1 is then achieved with or without information on the sparsity pattern and time required for each computation was recorded. For three numbers of selected animals (4000; 8000 and 12 000), A22 was computed using different pedigree extractions and the closeness of the resulting A22-1 to the inverse computed using the fully extracted pedigree was measured by an appropriate norm.
Results: The use of prior information on the sparsity of T-1 decreased the computing time for inversion by a factor of 1.73 on average. Computational issues and practical uses of the different algorithms were discussed. Cases involving more than 12 000 selected animals were considered. Inclusion of 10 generations was determined to be sufficient when computing A22.
Conclusions: Depending on the size and structure of the selected sub-population, gains in time to compute A22-1 are possible and these gains may increase as the number of selected animals increases. Given the sequential nature of most computational steps, the proposed algorithm can benefit from optimization and may be convenient for genomic evaluations.
Figures



Similar articles
-
A method to approximate the inverse of a part of the additive relationship matrix.J Anim Breed Genet. 2015 Jun;132(3):229-38. doi: 10.1111/jbg.12128. Epub 2015 Jan 6. J Anim Breed Genet. 2015. PMID: 25560252
-
Solving efficiently large single-step genomic best linear unbiased prediction models.J Anim Breed Genet. 2017 Jun;134(3):264-274. doi: 10.1111/jbg.12257. J Anim Breed Genet. 2017. PMID: 28508482
-
A recursive algorithm for decomposition and creation of the inverse of the genomic relationship matrix.J Dairy Sci. 2012 Oct;95(10):6093-102. doi: 10.3168/jds.2011-5249. Epub 2012 Aug 9. J Dairy Sci. 2012. PMID: 22884343
-
Is single-step genomic REML with the algorithm for proven and young more computationally efficient when less generations of data are present?J Anim Sci. 2022 May 1;100(5):skac082. doi: 10.1093/jas/skac082. J Anim Sci. 2022. PMID: 35289906 Free PMC article.
-
Symposium review: Single-step genomic evaluations in dairy cattle.J Dairy Sci. 2020 Jun;103(6):5314-5326. doi: 10.3168/jds.2019-17754. Epub 2020 Apr 22. J Dairy Sci. 2020. PMID: 32331883 Review.
Cited by
-
Extension of the reduced animal model to single-step methods.J Anim Sci. 2023 Jan 3;101:skac272. doi: 10.1093/jas/skac272. J Anim Sci. 2023. PMID: 36069946 Free PMC article.
References
-
- Wright S. Coefficients of inbreeding and relationship. American Nat. 1922;45:330–338. doi: 10.1086/279872. - DOI
-
- Henderson CR. In: Proceedings of the Animal Breeding and Genetics Symposium in Honour of JL Lush. American Society of Animal Science and American Dairy Science Association Champaign, editor. Champaign, IL: ; 1973. Sire evaluation and genetic trends; pp. 10–41.
-
- Gengler N, Nieuwhof G, Konstantinov K, Goddard M. Book of Abstracts of the 63rd Annual Meeting of the European Association of Animal Production: 27-31 August 2012. Bratislava Wageningen: Wageningen Academic Publishers; 2012. Alternative single-step type genomic prediction equations; p. 131.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

