Long-lived intracellular single-molecule fluorescence using electroporated molecules
- PMID: 24314075
- PMCID: PMC3853080
- DOI: 10.1016/j.bpj.2013.09.057
Long-lived intracellular single-molecule fluorescence using electroporated molecules
Abstract
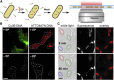
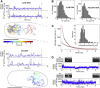
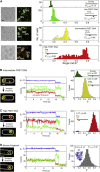
Studies of biomolecules in vivo are crucial to understand their function in a natural, biological context. One powerful approach involves fusing molecules of interest to fluorescent proteins to study their expression, localization, and action; however, the scope of such studies would be increased considerably by using organic fluorophores, which are smaller and more photostable than their fluorescent protein counterparts. Here, we describe a straightforward, versatile, and high-throughput method to internalize DNA fragments and proteins labeled with organic fluorophores into live Escherichia coli by employing electroporation. We studied the copy numbers, diffusion profiles, and structure of internalized molecules at the single-molecule level in vivo, and were able to extend single-molecule observation times by two orders of magnitude compared to green fluorescent protein, allowing continuous monitoring of molecular processes occurring from seconds to minutes. We also exploited the desirable properties of organic fluorophores to perform single-molecule Förster resonance energy transfer measurements in the cytoplasm of live bacteria, both for DNA and proteins. Finally, we demonstrate internalization of labeled proteins and DNA into yeast Saccharomyces cerevisiae, a model eukaryotic system. Our method should broaden the range of biological questions addressable in microbes by single-molecule fluorescence.
Copyright © 2013 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. - PubMed
-
- Yu J., Xiao J., Xie X.S. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. - PubMed
-
- Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

