Enhanced arbovirus surveillance with deep sequencing: Identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes
- PMID: 24314645
- PMCID: PMC3870893
- DOI: 10.1016/j.virol.2013.09.026
Enhanced arbovirus surveillance with deep sequencing: Identification of novel rhabdoviruses and bunyaviruses in Australian mosquitoes
Abstract
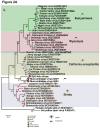
Viral metagenomics characterizes known and identifies unknown viruses based on sequence similarities to any previously sequenced viral genomes. A metagenomics approach was used to identify virus sequences in Australian mosquitoes causing cytopathic effects in inoculated mammalian cell cultures. Sequence comparisons revealed strains of Liao Ning virus (Reovirus, Seadornavirus), previously detected only in China, livestock-infecting Stretch Lagoon virus (Reovirus, Orbivirus), two novel dimarhabdoviruses, named Beaumont and North Creek viruses, and two novel orthobunyaviruses, named Murrumbidgee and Salt Ash viruses. The novel virus proteomes diverged by ≥ 50% relative to their closest previously genetically characterized viral relatives. Deep sequencing also generated genomes of Warrego and Wallal viruses, orbiviruses linked to kangaroo blindness, whose genomes had not been fully characterized. This study highlights viral metagenomics in concert with traditional arbovirus surveillance to characterize known and new arboviruses in field-collected mosquitoes. Follow-up epidemiological studies are required to determine whether the novel viruses infect humans.
Keywords: ABLV; AHSV; AINOV; AKAV; ALFV; ALMV; African horse sickness virus; Aino virus; Akabane virus; Alfuy virus; Almpiwar virus; Arbovirus surveillance; Australia; Australian bat lyssavirus; BANV; BASV; BEAUV; BEFV; BFV; BYSM; Banna virus; Barmah Forest virus; Bas-Congo virus; Beaumont virus; Bunyavirus; CCHFV; CHIKV; CHVV; CPV; Charleville virus; Coastal Plains virus; Crimean-Congo hemorrhagic fever virus; DAff sigma virus; DENV; DMel sigma virus; DOUV; DObs sigma virus; Deep sequencing; Douglas virus; Drosophila affinis sigma virus; Drosophila melanogaster sigma virus; Drosophila obscura sigma virus; EBLV; EHDV; EHV; EUBV; Edge Hill virus; Eubenangee virus; European bat lyssavirus; FPV; Facey's Paddock virus; GGV; Gan Gan virus; HDOOV; Humpty Doo virus; IHNV; JEV; Japanese encephalitis virus; KDV; KOKV; KUNV; Kadiporo virus; Kokobera virus; Kunjin virus; LEAV; LNV; LNYV; LYMoV; Leanyer virus; Liao Ning virus; MAPV; MFSV; MURBV; MVEV; Mapputta virus; Mosquito; Murray Valley encephalitis virus; Murrumbidgee virus; NCMV; NORCV; North Creek virus; Northern cereal mosaic virus; Novel virus; PEAV; PHSV; Peaton virus; Peruvian horse sickness virus; RRV; RVFV; Reovirus; Rhabdovirus; Rift Valley fever virus; Ross River virus; SASHV; SFTSV; SINV; SLOV; STRV; SVCV; Salt Ash virus; Sindbis virus; Stratford virus; Stretch Lagoon orbivirus; THIV; TIBV; TILV; TINV; TRUV; Thimiri virus; Tibrogargan virus; Tilligerry virus; Tinaroo virus; Trubanaman virus; VHSV; VSV; Virus discovery; WALV; WARV; WNV; Wallal virus; Warrego virus; West Nile virus; barley yellow striate mosaic; bovine ephemeral fever virus; chikungunya virus; dengue virus; epizootic hemorrhagic disease virus; infectious hematopoietic necrosis virus; lettuce necrotic yellows virus; lettuce yellow mottle virus; maize fine streak virus; severe fever with thrombocytopenia syndrome virus; spring viremia of carp virus; vesicular stomatitis virus; viral hemorrhagic septicemia virus.
© 2013 Elsevier Inc. All rights reserved.
Conflict of interest statement
None.
Figures



References
-
- Knope K, Whelan P, Smith D, Nicholson J, Moran R, Doggett S, Sly A, Hobby M, Wright P. Arboviral diseases and malaria in Australia, 2010–11: Annual report of the National Arbovirus and Malaria Advisory Committee. Commun Dis Intell Q Rep. 2013;37:E1–E20. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

