Modeling of the human rhinovirus C capsid suggests a novel topography with insights on receptor preference and immunogenicity
- PMID: 24314648
- PMCID: PMC3857591
- DOI: 10.1016/j.virol.2013.10.006
Modeling of the human rhinovirus C capsid suggests a novel topography with insights on receptor preference and immunogenicity
Abstract
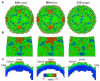
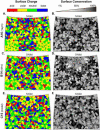
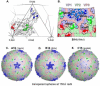
Features of human rhinovirus (RV)-C virions that allow them to use novel cell receptors and evade immune responses are unknown. Unlike the RV-A+B, these isolates cannot be propagated in typical culture systems or grown for structure studies. Comparative sequencing, I-TASSER, MODELLER, ROBETTA, and refined alignment techniques led to a structural approximation for C15 virions, based on the extensive, resolved RV-A+B datasets. The model predicts that all RV-C VP1 proteins are shorter by 21 residues relative to the RV-A, and 35 residues relative to the RV-B, effectively shaving the RV 5-fold plateau from the particle. There are major alterations in VP1 neutralizing epitopes and the structural determinants for ICAM-1 and LDLR receptors. The VP2 and VP3 elements are similar among all RV, but the loss of sequence "words" contributing Nim1ab has increased the apparent selective pressure among the RV-C to fix mutations elsewhere in the VP1, creating a possible compensatory epitope.
Keywords: Capsid structure; I-Tasser; Immunogenicity; Model; Receptor binding; Rhinovirus; Rhinovirus C.
© 2013 Elsevier Inc. All rights reserved.
Figures



References
-
- Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas T, Printz M, Lee W, Shult P, Reisdorf E, Carlson-Dakes K, Salazar L, Dasilva D, Tisler C, Gern J, Lemanske R. Wheezing Rhinovirus Illnesses in Early Life Predict Asthma Development in High-Risk Children. Am J Respir Crit Care Med. 2008;178:667–672. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

