Gene duplication and phylogeography of North American members of the Hart Park serogroup of avian rhabdoviruses
- PMID: 24314659
- PMCID: PMC3873333
- DOI: 10.1016/j.virol.2013.10.024
Gene duplication and phylogeography of North American members of the Hart Park serogroup of avian rhabdoviruses
Abstract
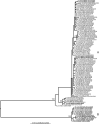
Flanders virus (FLAV) and Hart Park virus (HPV) are rhabdoviruses that circulate in mosquito-bird cycles in the eastern and western United States, respectively, and constitute the only two North American representatives of the Hart Park serogroup. Previously, it was suggested that FLAV is unique among the rhabdoviruses in that it contains two pseudogenes located between the P and M genes, while the cognate sequence for HPV has been lacking. Herein, we demonstrate that FLAV and HPV do not contain pseudogenes in this region, but encode three small functional proteins designated as U1-U3 that apparently arose by gene duplication. To further investigate the U1-U3 region, we conducted the first large-scale evolutionary analysis of a member of the Hart Park serogroup by analyzing over 100 spatially and temporally distinct FLAV isolates. Our phylogeographic analysis demonstrates that although FLAV appears to be slowly evolving, phylogenetically divergent lineages co-circulate sympatrically.
Keywords: Bird-associated arbovirus; Coupled translation; Flanders virus; Gene duplication; Hart Park serogroup; Hart Park virus; Rhabdovirus; SH protein; U1–U3 proteins.
© 2013 Published by Elsevier Inc.
Figures




References
-
- Bose S., Mathur M., Bates P., Joshi N., Banerjee A.K. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 2003;84:1687–1699. - PubMed
-
- Bourhy H., Cowley J.A., Larrous F., Holmes E.C., Walker P.J. Phylogenetic relationships among rhabdoviruses inferred using the L polymerase gene. J. Gen. Virol. 2005;86:2849–2858. - PubMed
-
- Bourhy H., Gubala A.J., Weir R.P., Boyle D.B. Animal Rhabdoviruses. In: Mahy B.W.J., Van Regenmortel M.H.V., editors. Encyclopedia of Virology. third ed. Elsevier Academic Press; San Diego, California: 2008. pp. 111–121.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

