Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing
- PMID: 24315099
- PMCID: PMC3902092
- DOI: 10.1016/j.cell.2013.10.045
Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing
Abstract
Bacteria that cause disease rely on their ability to counteract and overcome host defenses. Here, we present a genome-scale study of Mycobacterium tuberculosis (Mtb) that uncovers the bacterial determinants of surviving host immunity, sets of genes we term "counteractomes." Through this analysis, we found that CD4 T cells attempt to contain Mtb growth by starving it of tryptophan--a mechanism that successfully limits infections by Chlamydia and Leishmania, natural tryptophan auxotrophs. Mtb, however, can synthesize tryptophan under stress conditions, and thus, starvation fails as an Mtb-killing mechanism. We then identify a small-molecule inhibitor of Mtb tryptophan synthesis, which converts Mtb into a tryptophan auxotroph and restores the efficacy of a failed host defense. Together, our findings demonstrate that the Mtb immune counteractomes serve as probes of host immunity, uncovering immune-mediated stresses that can be leveraged for therapeutic discovery.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

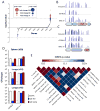




Comment in
-
Trp'ing tuberculosis.Cell. 2013 Dec 5;155(6):1209-10. doi: 10.1016/j.cell.2013.11.015. Cell. 2013. PMID: 24315090
References
-
- Alberati-Giani D, Malherbe P, Ricciardi-Castagnoli P, Kohler C, Denis-Donini S, Cesura A. Differential Regulation of Indoleamine 2,3-Dioxygenase Expression by Nitric Oxide and Inflammatory Mediators IFN-gamma-Activated Murine Macrophages and Microglial Cells. Journal of immunology (Baltimore, Md : 1950) 1997;159:419–426. - PubMed
-
- Baban B, Chandler P, McCool D, Marshall B, Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase expression is restricted to fetal trophoblast giant cells during murine gestation and is maternal genome specific. Journal of reproductive immunology. 2004;61:67–77. - PubMed
-
- Baker TI, Crawford IP. Anthranilate synthetase: partical purification and some kinetic studies on the enzyme from Eschericia coli. Journal of Biological Chemistry. 1966;241:5577–5584. - PubMed
-
- Bauerle R, Hess J, French S. Anthranilate synthase-anthranilate phosphoribosyl transferase complex and subunits of Salmonella typhimurium. Methods Enzymol. 1987;142:366–386. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

