Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland
- PMID: 24315934
- PMCID: PMC3944000
- DOI: 10.1016/j.chembiol.2013.10.015
Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland
Abstract
Nature combines existing biochemical building blocks, at times with subtlety of purpose. RNA modifications are a prime example of this, where standard RNA nucleosides are decorated with chemical groups and building blocks that we recall from our basic biochemistry lectures. The result: a wealth of chemical diversity whose full biological relevance has remained elusive despite being public knowledge for some time. Here, we highlight several modifications that, because of their chemical intricacy, rely on seemingly unrelated pathways to provide cofactors for their synthesis. Besides their immediate role in affecting RNA function, modifications may act as sensors and transducers of information that connect a cell's metabolic state to its translational output, carefully orchestrating a delicate balance between metabolic rate and protein synthesis at a system's level.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures

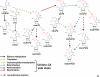

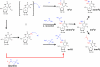

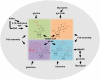
References
-
- Armengod ME, Moukadiri I, Prado S, Ruiz-Partida R, Benitez-Paez A, Villarroya M, Lomas R, Garzon MJ, Martinez-Zamora A, Meseguer S, Navarro-Gonzalez C. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie. 2012;94:1510–1520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

