Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis
- PMID: 24316671
- PMCID: PMC8369836
- DOI: 10.1038/ncb2883
Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis
Erratum in
- Nat Cell Biol. 2014 Feb;16(2):200
Abstract
The mixed lineage kinase domain-like protein (MLKL) has recently been identified as a key RIP3 (receptor interacting protein 3) downstream component of tumour necrosis factor (TNF)-induced necroptosis. MLKL is phosphorylated by RIP3 and is recruited to the necrosome through its interaction with RIP3. However, it is still unknown how MLKL mediates TNF-induced necroptosis. Here, we report that MLKL forms a homotrimer through its amino-terminal coiled-coil domain and locates to the cell plasma membrane during TNF-induced necroptosis. By generating different MLKL mutants, we demonstrated that the plasma membrane localization of trimerized MLKL is critical for mediating necroptosis. Importantly, we found that the membrane localization of MLKL is essential for Ca(2+) influx, which is an early event of TNF-induced necroptosis. Furthermore, we identified that TRPM7 (transient receptor potential melastatin related 7) is a MLKL downstream target for the mediation of Ca(2+) influx and TNF-induced necroptosis. Hence, our study reveals a crucial mechanism of MLKL-mediated TNF-induced necroptosis.
Figures

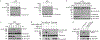


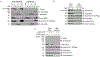

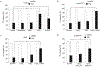

References
-
- Chen G & Goeddel DV TNF-R1 signaling: a beautiful pathway. Science 296, 1634–1635 (2002). - PubMed
-
- Ashkenazi A & Dixit VM Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol 11, 255–260 (1999). - PubMed
-
- Vandenabeele P, Galluzzi L, Vanden Berghe T & Kroemer G Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. Cell Biol 11, 700–714 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

