Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1
- PMID: 24316673
- PMCID: PMC3876036
- DOI: 10.1038/ncb2886
Autophagy variation within a cell population determines cell fate through selective degradation of Fap-1
Abstract
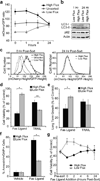
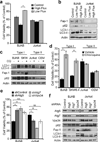
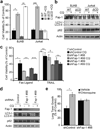
Autophagy regulates cell death both positively and negatively, but the molecular basis for this paradox remains inadequately characterized. We demonstrate here that transient cell-to-cell variations in autophagy can promote either cell death or survival depending on the stimulus and cell type. By separating cells with high and low basal autophagy using flow cytometry, we demonstrate that autophagy determines which cells live or die in response to death receptor activation. We have determined that selective autophagic degradation of the phosphatase Fap-1 promotes Fas apoptosis in Type I cells, which do not require mitochondrial permeabilization for efficient apoptosis. Conversely, autophagy inhibits apoptosis in Type II cells (which require mitochondrial involvement) or on treatment with TRAIL in either Type I or II cells. These data illustrate that differences in autophagy in a cell population determine cell fate in a stimulus- and cell-type-specific manner. This example of selective autophagy of an apoptosis regulator may represent a general mechanism for context-specific regulation of cell fate by autophagy.
Figures



Comment in
-
Autophagy chews Fap to promote apoptosis.Nat Cell Biol. 2014 Jan;16(1):23-5. doi: 10.1038/ncb2899. Nat Cell Biol. 2014. PMID: 24366033
References
-
- Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

