MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis
- PMID: 24316975
- PMCID: PMC3871244
- DOI: 10.1172/JCI71180
MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis
Abstract
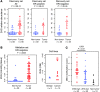
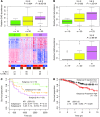
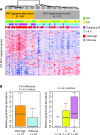
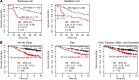
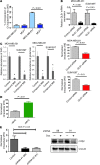
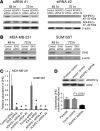
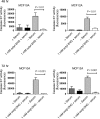
Metabolic profiling of cancer cells has recently been established as a promising tool for the development of therapies and identification of cancer biomarkers. Here we characterized the metabolomic profile of human breast tumors and uncovered intrinsic metabolite signatures in these tumors using an untargeted discovery approach and validation of key metabolites. The oncometabolite 2-hydroxyglutarate (2HG) accumulated at high levels in a subset of tumors and human breast cancer cell lines. We discovered an association between increased 2HG levels and MYC pathway activation in breast cancer, and further corroborated this relationship using MYC overexpression and knockdown in human mammary epithelial and breast cancer cells. Further analyses revealed globally increased DNA methylation in 2HG-high tumors and identified a tumor subtype with high tissue 2HG and a distinct DNA methylation pattern that was associated with poor prognosis and occurred with higher frequency in African-American patients. Tumors of this subtype had a stem cell-like transcriptional signature and tended to overexpress glutaminase, suggestive of a functional relationship between glutamine and 2HG metabolism in breast cancer. Accordingly, 13C-labeled glutamine was incorporated into 2HG in cells with aberrant 2HG accumulation, whereas pharmacologic and siRNA-mediated glutaminase inhibition reduced 2HG levels. Our findings implicate 2HG as a candidate breast cancer oncometabolite associated with MYC activation and poor prognosis.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

