Structural features and mechanism of translocation of non-LTR retrotransposons in Candida albicans
- PMID: 24317340
- PMCID: PMC3956500
- DOI: 10.4161/viru.27278
Structural features and mechanism of translocation of non-LTR retrotransposons in Candida albicans
Abstract
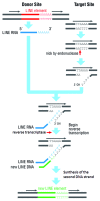
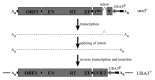
A number of abundant mobile genetic elements called retrotransposons reverse transcribe RNA to generate DNA for insertion into eukaryotic genomes. Non-long-terminal repeat (non-LTR) retrotransposons represent a major class of retrotransposons, and transposons that move by target-primed reverse transcription lack LTRs characteristic of retroviruses and retroviral-like transposons. Yeast model systems in Candida albicans and Saccharomyces cerevisiae have been developed for the study of non-LTR retrotransposons. Non-LTR retrotransposons are divided into LINEs (long interspersed nuclear elements), SINEs (short interspersed nuclear elements), and SVA (SINE, VNTR, and Alu). LINE-1 elements have been described in fungi, and several families called Zorro elements have been detected from C. albicans. They are all members of L1 clades. Through a mechanism named target-primed reverse transcription (TPRT), LINEs translocate the new copy into the target site to initiate DNA synthesis primed by the 3' OH of the broken strand. In this article, we describe some advances in the research on structural features and origin of non-LTR retrotransposons in C. albicans, and discuss mechanisms underlying their reverse transcription and integration of the donor copy into the target site.
Keywords: Candida albicans; LINE; Zorro; non-LTR retrotransposons; target-primed reverse transcription (TPRT).
Figures



Similar articles
-
The structure and retrotransposition mechanism of LTR-retrotransposons in the asexual yeast Candida albicans.Virulence. 2014 Aug 15;5(6):655-64. doi: 10.4161/viru.32180. Epub 2014 Aug 7. Virulence. 2014. PMID: 25101670 Free PMC article. Review.
-
L1-like non-LTR retrotransposons in the yeast Candida albicans.Curr Genet. 2001 Apr;39(2):83-91. doi: 10.1007/s002940000181. Curr Genet. 2001. PMID: 11405100
-
[Non-LTR retrotransposons: LINEs and SINEs in plant genome].Yi Chuan. 2006 Jun;28(6):731-6. Yi Chuan. 2006. PMID: 16818439 Review. Chinese.
-
The diversity of retrotransposons and the properties of their reverse transcriptases.Virus Res. 2008 Jun;134(1-2):221-34. doi: 10.1016/j.virusres.2007.12.010. Epub 2008 Feb 7. Virus Res. 2008. PMID: 18261821 Free PMC article. Review.
-
LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant genomes.Plant Mol Biol. 1999 Aug;40(6):903-10. doi: 10.1023/a:1006212929794. Plant Mol Biol. 1999. PMID: 10527415 Review.
Cited by
-
Genome-wide analysis of heat stress-stimulated transposon mobility in the human fungal pathogen Cryptococcus deneoformans.Proc Natl Acad Sci U S A. 2023 Jan 24;120(4):e2209831120. doi: 10.1073/pnas.2209831120. Epub 2023 Jan 20. Proc Natl Acad Sci U S A. 2023. PMID: 36669112 Free PMC article.
-
Advances in Molecular Tools and In Vivo Models for the Study of Human Fungal Pathogenesis.Microorganisms. 2020 May 26;8(6):803. doi: 10.3390/microorganisms8060803. Microorganisms. 2020. PMID: 32466582 Free PMC article. Review.
References
-
- Odds FC. A Review and Bibliography. Candida and Candidosis, London: Bailliere Tindall; 1988.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
