Transcription-generated torsional stress destabilizes nucleosomes
- PMID: 24317489
- PMCID: PMC3947361
- DOI: 10.1038/nsmb.2723
Transcription-generated torsional stress destabilizes nucleosomes
Abstract
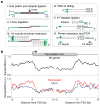
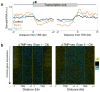
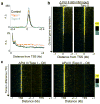
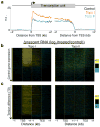
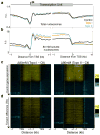
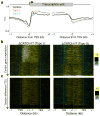
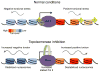
As RNA polymerase II (Pol II) transcribes a gene, it encounters an array of well-ordered nucleosomes. How it traverses through this array in vivo remains unresolved. One model proposes that torsional stress generated during transcription destabilizes nucleosomes ahead of Pol II. Here, we describe a method for high-resolution mapping of underwound DNA, using next-generation sequencing, and show that torsion is correlated with gene expression in Drosophila melanogaster cells. Accumulation of torsional stress, through topoisomerase inhibition, results in increased Pol II at transcription start sites. Whereas topoisomerase I inhibition results in increased nascent RNA transcripts, topoisomerase II inhibition causes little change. Despite the different effects on Pol II elongation, topoisomerase inhibition results in increased nucleosome turnover and salt solubility within gene bodies, thus suggesting that the elongation-independent effects of torsional stress on nucleosome dynamics contributes to the destabilization of nucleosomes.
Figures
References
-
- Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. - PubMed
-
- Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. - PubMed
-
- Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

