Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions
- PMID: 24317492
- PMCID: PMC3947387
- DOI: 10.1038/nsmb.2718
Overlapping chromatin-remodeling systems collaborate genome wide at dynamic chromatin transitions
Abstract
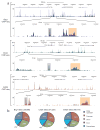
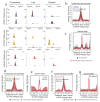
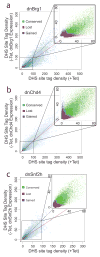
ATP-dependent chromatin remodeling is an essential process required for the dynamic organization of chromatin structure. Here we describe the genome-wide location and activity of three remodeler proteins with diverse physiological functions in the mouse genome: Brg1, Chd4 and Snf2h. The localization patterns of all three proteins substantially overlap with one another and with regions of accessible chromatin. Furthermore, using inducible mutant variants, we demonstrate that the catalytic activity of these proteins contributes to the remodeling of chromatin genome wide and that each of these remodelers can independently regulate chromatin reorganization at distinct sites. Many regions require the activity of more than one remodeler to regulate accessibility. These findings provide a dynamic view of chromatin organization and highlight the differential contributions of remodelers to chromatin maintenance in higher eukaryotes.
Figures




References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

