RGS6, but not RGS4, is the dominant regulator of G protein signaling (RGS) modulator of the parasympathetic regulation of mouse heart rate
- PMID: 24318880
- PMCID: PMC3900986
- DOI: 10.1074/jbc.M113.520742
RGS6, but not RGS4, is the dominant regulator of G protein signaling (RGS) modulator of the parasympathetic regulation of mouse heart rate
Abstract
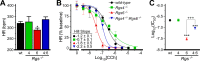
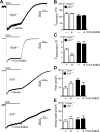
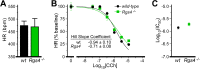
Parasympathetic activity decreases heart rate (HR) by inhibiting pacemaker cells in the sinoatrial node (SAN). Dysregulation of parasympathetic influence has been linked to sinus node dysfunction and arrhythmia. RGS (regulator of G protein signaling) proteins are negative modulators of the parasympathetic regulation of HR and the prototypical M2 muscarinic receptor (M2R)-dependent signaling pathway in the SAN that involves the muscarinic-gated atrial K(+) channel IKACh. Both RGS4 and RGS6-Gβ5 have been implicated in these processes. Here, we used Rgs4(-/-), Rgs6(-/-), and Rgs4(-/-):Rgs6(-/-) mice to compare the relative influence of RGS4 and RGS6 on parasympathetic regulation of HR and M2R-IKACh-dependent signaling in the SAN. In retrogradely perfused hearts, ablation of RGS6, but not RGS4, correlated with decreased resting HR, increased heart rate variability, and enhanced sensitivity to the negative chronotropic effects of the muscarinic agonist carbachol. Similarly, loss of RGS6, but not RGS4, correlated with enhanced sensitivity of the M2R-IKACh signaling pathway in SAN cells to carbachol and a significant slowing of M2R-IKACh deactivation rate. Surprisingly, concurrent genetic ablation of RGS4 partially rescued some deficits observed in Rgs6(-/-) mice. These findings, together with those from an acute pharmacologic approach in SAN cells from Rgs6(-/-) and Gβ5(-/-) mice, suggest that the partial rescue of phenotypes in Rgs4(-/-):Rgs6(-/-) mice is attributable to another R7 RGS protein whose influence on M2R-IKACh signaling is masked by RGS4. Thus, RGS6-Gβ5, but not RGS4, is the primary RGS modulator of parasympathetic HR regulation and SAN M2R-IKACh signaling in mice.
Keywords: G Proteins; GIRK/Kir3; Gene Knockout; Heart; Muscarinic; Patch Clamp Electrophysiology; Potassium Channels; RGS Proteins; Sinoatrial Node.
Figures


Similar articles
-
GPCR-dependent biasing of GIRK channel signaling dynamics by RGS6 in mouse sinoatrial nodal cells.Proc Natl Acad Sci U S A. 2020 Jun 23;117(25):14522-14531. doi: 10.1073/pnas.2001270117. Epub 2020 Jun 8. Proc Natl Acad Sci U S A. 2020. PMID: 32513692 Free PMC article.
-
RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node.Circ Res. 2008 Aug 29;103(5):527-35. doi: 10.1161/CIRCRESAHA.108.180984. Epub 2008 Jul 24. Circ Res. 2008. PMID: 18658048
-
Essential role of the m2R-RGS6-IKACh pathway in controlling intrinsic heart rate variability.PLoS One. 2013 Oct 29;8(10):e76973. doi: 10.1371/journal.pone.0076973. eCollection 2013. PLoS One. 2013. PMID: 24204714 Free PMC article.
-
Structure, function, and localization of Gβ5-RGS complexes.Prog Mol Biol Transl Sci. 2009;86:157-203. doi: 10.1016/S1877-1173(09)86006-7. Epub 2009 Oct 7. Prog Mol Biol Transl Sci. 2009. PMID: 20374716 Free PMC article. Review.
-
RGS6 as a Novel Therapeutic Target in CNS Diseases and Cancer.AAPS J. 2016 May;18(3):560-72. doi: 10.1208/s12248-016-9899-9. Epub 2016 Mar 22. AAPS J. 2016. PMID: 27002730 Free PMC article. Review.
Cited by
-
Rapid and accurate multi-phenotype imputation for millions of individuals.Nat Commun. 2025 Jan 4;16(1):387. doi: 10.1038/s41467-024-55496-0. Nat Commun. 2025. PMID: 39755672 Free PMC article.
-
Protein Profiling of RGS6, a Pleiotropic Gene Implicated in Numerous Neuropsychiatric Disorders, Reveals Multi-Isoformic Expression and a Novel Brain-Specific Isoform.eNeuro. 2022 Jan 18;9(1):ENEURO.0379-21.2021. doi: 10.1523/ENEURO.0379-21.2021. Print 2022 Jan-Feb. eNeuro. 2022. PMID: 34880111 Free PMC article.
-
VU0810464, a non-urea G protein-gated inwardly rectifying K+ (Kir 3/GIRK) channel activator, exhibits enhanced selectivity for neuronal Kir 3 channels and reduces stress-induced hyperthermia in mice.Br J Pharmacol. 2019 Jul;176(13):2238-2249. doi: 10.1111/bph.14671. Epub 2019 May 30. Br J Pharmacol. 2019. PMID: 30924523 Free PMC article.
-
Genetics and the heart rate response to exercise.Cell Mol Life Sci. 2019 Jun;76(12):2391-2409. doi: 10.1007/s00018-019-03079-4. Epub 2019 Mar 27. Cell Mol Life Sci. 2019. PMID: 30919020 Free PMC article. Review.
-
Genetic Analysis of Rare Human Variants of Regulators of G Protein Signaling Proteins and Their Role in Human Physiology and Disease.Pharmacol Rev. 2018 Jul;70(3):446-474. doi: 10.1124/pr.117.015354. Pharmacol Rev. 2018. PMID: 29871944 Free PMC article. Review.
References
-
- Mangoni M. E., Nargeot J. (2008) Genesis and regulation of the heart automaticity. Physiol. Rev. 88, 919–982 - PubMed
-
- DiFrancesco D. (2010) The role of the funny current in pacemaker activity. Circ. Res. 106, 434–446 - PubMed
-
- Dobrzynski H., Boyett M. R., Anderson R. H. (2007) New insights into pacemaker activity: promoting understanding of sick sinus syndrome. Circulation 115, 1921–1932 - PubMed
-
- Fu Y., Huang X., Piao L., Lopatin A. N., Neubig R. R. (2007) Endogenous RGS proteins modulate SA and AV nodal functions in isolated heart: implications for sick sinus syndrome and AV block. Am. J. Physiol. Heart Circ. Physiol. 292, H2532–H2539 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

