Roles of rpoS-activating small RNAs in pathways leading to acid resistance of Escherichia coli
- PMID: 24319011
- PMCID: PMC3937726
- DOI: 10.1002/mbo3.143
Roles of rpoS-activating small RNAs in pathways leading to acid resistance of Escherichia coli
Abstract
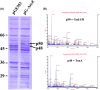
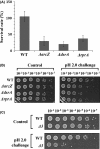
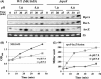
Escherichia coli and related enteric bacteria can survive under extreme acid stress condition at least for several hours. RpoS is a key factor for acid stress management in many enterobacteria. Although three rpoS-activating sRNAs, DsrA, RprA, and ArcZ, have been identified in E. coli, it remains unclear how these small RNA molecules participate in pathways leading to acid resistance (AR). Here, we showed that overexpression of ArcZ, DsrA, or RprA enhances AR in a RpoS-dependent manner. Mutant strains with deletion of any of three sRNA genes showed lowered AR, and deleting all three sRNA genes led to more severe defects in protecting against acid stress. Overexpression of any of the three sRNAs fully rescued the acid tolerance defects of the mutant strain lacking all three genes, suggesting that all three sRNAs perform the same function in activating RpoS required for AR. Notably, acid stress led to the induction of DsrA and RprA but not ArcZ.
Keywords: Acid resistance; Escherichia coli; RpoS; small noncoding RNA..
© 2013 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Figures






References
-
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. - PubMed
-
- Bordi C, Theraulaz L, Mejean V, Jourlin-Castelli C. Anticipating an alkaline stress through the Tor phosphorelay system in Escherichia coli. Mol. Microbiol. 2003;48:211–223. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

