The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events
- PMID: 24319082
- PMCID: PMC3912096
- DOI: 10.1104/pp.113.231928
The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events
Abstract
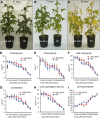
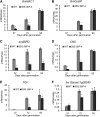
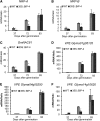
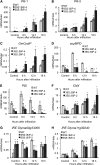
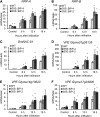
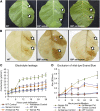
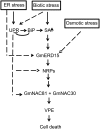
The binding protein (BiP) has been demonstrated to participate in innate immunity and attenuate endoplasmic reticulum- and osmotic stress-induced cell death. Here, we employed transgenic plants with manipulated levels of BiP to assess whether BiP also controlled developmental and hypersensitive programmed cell death (PCD). Under normal conditions, the BiP-induced transcriptome revealed a robust down-regulation of developmental PCD genes and an up-regulation of the genes involved in hypersensitive PCD triggered by nonhost-pathogen interactions. Accordingly, the BiP-overexpressing line displayed delayed leaf senescence under normal conditions and accelerated hypersensitive response triggered by Pseudomonas syringae pv tomato in soybean (Glycine max) and tobacco (Nicotiana tabacum), as monitored by measuring hallmarks of PCD in plants. The BiP-mediated delay of leaf senescence correlated with the attenuation of N-rich protein (NRP)-mediated cell death signaling and the inhibition of the senescence-associated activation of the unfolded protein response (UPR). By contrast, under biological activation of salicylic acid (SA) signaling and hypersensitive PCD, BiP overexpression further induced NRP-mediated cell death signaling and antagonistically inhibited the UPR. Thus, the SA-mediated induction of NRP cell death signaling occurs via a pathway distinct from UPR. Our data indicate that during the hypersensitive PCD, BiP positively regulates the NRP cell death signaling through a yet undefined mechanism that is activated by SA signaling and related to ER functioning. By contrast, BiP's negative regulation of leaf senescence may be linked to its capacity to attenuate the UPR activation and NRP cell death signaling. Therefore, BiP can function either as a negative or positive modulator of PCD events.
Figures



References
-
- Bogdanove AJ, Kim JF, Wei ZM, Kolchinsky P, Charkowski AO, Conlin AK, Collmer A, Beer SV. (1998) Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc Natl Acad Sci USA 95: 1325–1330 - PMC - PubMed
-
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
-
- Brandizzi F, Hanton SL, DaSilva LLP, Boevink P, Evans D, Oparka K, Denecke J, Hawes C. (2003) ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant J 34: 269–281 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

