Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients
- PMID: 24323028
- PMCID: PMC4062710
- DOI: 10.1200/JCO.2013.50.8556
Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients
Abstract
Purpose: Lung squamous cell carcinoma (SCC) is the second most prevalent type of lung cancer. Currently, no targeted therapeutics are approved for treatment of this cancer, largely because of a lack of systematic understanding of the molecular pathogenesis of the disease. To identify therapeutic targets and perform comparative analyses of lung SCC, we probed somatic genome alterations of lung SCC by using samples from Korean patients.
Patients and methods: We performed whole-exome sequencing of DNA from 104 lung SCC samples from Korean patients and matched normal DNA. In addition, copy-number analysis and transcriptome analysis were conducted for a subset of these samples. Clinical association with cancer-specific somatic alterations was investigated.
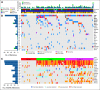
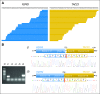
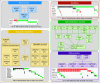
Results: This cancer cohort is characterized by a high mutational burden with an average of 261 somatic exonic mutations per tumor and a mutational spectrum showing a signature of exposure to cigarette smoke. Seven genes demonstrated statistical enrichment for mutation: TP53, RB1, PTEN, NFE2L2, KEAP1, MLL2, and PIK3CA). Comparative analysis between Korean and North American lung SCC samples demonstrated a similar spectrum of alterations in these two populations in contrast to the differences seen in lung adenocarcinoma. We also uncovered recurrent occurrence of therapeutically actionable FGFR3-TACC3 fusion in lung SCC.
Conclusion: These findings provide new steps toward the identification of genomic target candidates for precision medicine in lung SCC, a disease with significant unmet medical needs.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures
References
-
- World Health Organization. Cancer Fact Sheet No. 297. http://www.who.int/mediacentre/factsheets/fs297/en/
-
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. - PubMed
-
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

