The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes
- PMID: 24323502
- PMCID: PMC3904712
- DOI: 10.1093/jxb/ert399
The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes
Abstract
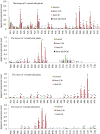
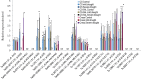
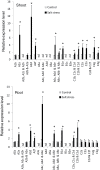
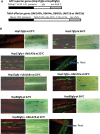
Heat shock factors (Hsfs) play a central regulatory role in acquired thermotolerance. To understand the role of the major molecular players in wheat adaptation to heat stress, the Hsf family was investigated in Triticum aestivum. Bioinformatic and phylogenetic analyses identified 56 TaHsf members, which are classified into A, B, and C classes. Many TaHsfs were constitutively expressed. Subclass A6 members were predominantly expressed in the endosperm under non-stress conditions. Upon heat stress, the transcript levels of A2 and A6 members became the dominant Hsfs, suggesting an important regulatory role during heat stress. Many TaHsfA members as well as B1, C1, and C2 members were also up-regulated during drought and salt stresses. The heat-induced expression profiles of many heat shock protein (Hsp) genes were paralleled by those of A2 and A6 members. Transactivation analysis revealed that in addition to TaHsfA members (A2b and A4e), overexpression of TaHsfC2a activated expression of TaHsp promoter-driven reporter genes under non-stress conditions, while TaHsfB1b and TaHsfC1b did not. Functional heat shock elements (HSEs) interacting with TaHsfA2b were identified in four TaHsp promoters. Promoter mutagenesis analysis demonstrated that an atypical HSE (GAACATTTTGGAA) in the TaHsp17 promoter is functional for heat-inducible expression and transactivation by Hsf proteins. The transactivation of Hsp promoter-driven reporter genes by TaHsfC2a also relied on the presence of HSE. An activation motif in the C-terminal domain of TaHsfC2a was identified by amino residue substitution analysis. These data demonstrate the role of HsfA and HsfC2 in regulation of Hsp genes in wheat.
Keywords: Gene expression; gene regulation; heat shock factors; heat shock proteins; heat stress; transcription factors; wheat..
Figures





References
-
- Almoguera C, Rojas A, Díaz-Martín J, Prieto-Dapena P, Carranco R, Jordano J. 2002. A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. Journal of Biological Chemistry 277, 43866–43872 - PubMed
-
- Bharti K, von Koskull-Doring P, Bharti S, Kumar P, Tintschl-Korbitzer A, Treuter E, Nover L. 2004. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. The Plant Cell 16, 1521–1535 - PMC - PubMed
-
- Busch W, Wunderlich M, Schöffl F. 2005. Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana . The Plant Journal 41, 1–14 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

