How do I kill thee? Let me count the ways: p53 regulates PARP-1 dependent necrosis
- PMID: 24323920
- PMCID: PMC3914234
- DOI: 10.1002/bies.201300117
How do I kill thee? Let me count the ways: p53 regulates PARP-1 dependent necrosis
Abstract
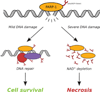
Understanding the impact of the p53 tumor suppressor pathway on the regulation of genome integrity, cancer development, and cancer treatment has intrigued scientists and clinicians for decades. It appears that the p53 pathway is a central node for nearly all cell stress responses, including: gene expression, DNA repair, cell cycle arrest, metabolic adjustments, apoptosis, and senescence. In the past decade, it has become increasingly clear that p53 function is directly regulated by poly(ADP-ribose) polymerase-1 (PARP-1), a nuclear enzyme involved in DNA repair signaling. Here, we will discuss the impact of PARP-1 on p53 function, along with a recently described novel role for the reciprocal regulation of p53 regulated, PARP-1 dependent necrosis following DNA damage.
Keywords: PARP-1; ROS; apoptosis; necrosis; p53.
© 2014 WILEY Periodicals, Inc.
Figures


References
-
- Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. - PubMed
-
- Linzer DI, Levine AJ. Characterization of a 54 K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. - PubMed
-
- Linzer DI, Maltzman W, Levine AJ. The SV40 A gene product is required for the production of a 54,000MW cellular tumor antigen. Virology. 1979;98:308–318. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

