Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors
- PMID: 24324157
- PMCID: PMC3876203
- DOI: 10.1073/pnas.1319097110
Visualization of repetitive DNA sequences in human chromosomes with transcription activator-like effectors
Erratum in
- Proc Natl Acad Sci U S A. 2014 Jan 21;111(3):1222
Abstract
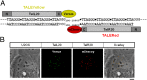
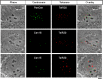
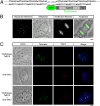
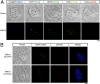
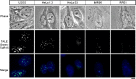
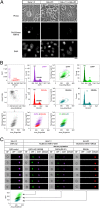
We describe a transcription activator-like effector (TALE)-based strategy, termed "TALEColor," for labeling specific repetitive DNA sequences in human chromosomes. We designed TALEs for the human telomeric repeat and fused them with any of numerous fluorescent proteins (FPs). Expression of these TALE-telomere-FP fusion proteins in human osteosarcoma's (U2OS) cells resulted in bright signals coincident with telomeres. We also designed TALEs for centromeric sequences unique to certain chromosomes, enabling us to localize specific human chromosomes in live cells. Meanwhile we generated TALE-FPs in vitro and used them as probes to detect telomeres in fixed cells. Using human cells with different average telomere lengths, we found that the TALEColor signals correlated positively with telomere length. In addition, suspension cells were followed by imaging flow cytometry to resolve cell populations with differing telomere lengths. These methods may have significant potential both for basic chromosome and genome research as well as in clinical applications.
Conflict of interest statement
Conflict of interest statement: H.M. is an inventor on a US patent application filed by the University of Massachusetts.
Figures
Comment in
-
Repeated TALEs: visualizing DNA sequence localization and chromosome dynamics in live cells.Nucleus. 2014 Jan-Feb;5(1):28-31. doi: 10.4161/nucl.28143. Epub 2014 Feb 10. Nucleus. 2014. PMID: 24637394 Free PMC article.
References
-
- Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. - PubMed
-
- Baker M. Gene-editing nucleases. Nat Methods. 2012;9(1):23–26. - PubMed
-
- Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

