Glycation accelerates fibrillization of the amyloidogenic W7FW14F apomyoglobin
- PMID: 24324625
- PMCID: PMC3851467
- DOI: 10.1371/journal.pone.0080768
Glycation accelerates fibrillization of the amyloidogenic W7FW14F apomyoglobin
Retraction in
-
Retraction: Glycation Accelerates Fibrillization of the Amyloidogenic W7FW14F Apomyoglobin.PLoS One. 2019 Jun 6;14(6):e0218284. doi: 10.1371/journal.pone.0218284. eCollection 2019. PLoS One. 2019. PMID: 31170266 Free PMC article. No abstract available.
Abstract
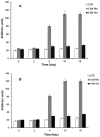
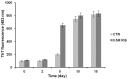
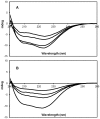
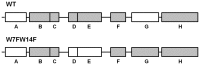
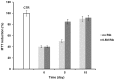
Neurodegenerative diseases are associated with misfolding and deposition of specific proteins, either intra or extracellularly in the nervous system. Advanced glycation end products (AGEs) originate from different molecular species that become glycated after exposure to sugars. Several proteins implicated in neurodegenerative diseases have been found to be glycated in vivo and the extent of glycation is related to the pathologies of the patients. Although it is now accepted that there is a direct correlation between AGEs formation and the development of neurodegenerative diseases, several questions still remain unanswered: whether glycation is the triggering event or just an additional factor acting on the aggregation pathway. To this concern, in the present study we have investigated the effect of glycation on the aggregation pathway of the amyloidogenic W7FW14F apomyoglobin. Although this protein has not been related to any amyloid disease, it represents a good model to resemble proteins that intrinsically evolve toward the formation of amyloid aggregates in physiological conditions. We show that D-ribose, but not D-glucose, rapidly induces the W7FW14F apomyoglobin to generate AGEs in a time-dependent manner and protein ribosylation is likely to involve lysine residues on the polypeptide chain. Ribosylation of the W7FW14F apomyoglobin strongly affects its aggregation kinetics producing amyloid fibrils within few days. Cytotoxicity of the glycated aggregates has also been tested using a cell viability assay. We propose that ribosylation in the W7FW14F apomyoglobin induces the formation of a cross-link that strongly reduces the flexibility of the H helix and/or induce a conformational change that favor fibril formation. These results open new perspectives for AGEs biological role as they can be considered not only a triggering factor in amyloidosis but also a player in later stages of the aggregation process.
Conflict of interest statement
Figures


References
-
- Dobson CM (2003) Protein folding and misfolding. Nature 426: 884–890. - PubMed
-
- Luheshi LM, Dobson CM (2009) Bridging the gap: from protein misfolding to protein mis-folding diseases. FEBS Letters 583: 2581–2586. - PubMed
-
- Serpell LC, Sunde M, Benson MD, Tennent GA, Pepys MB, et al. (2000) The protofilament substructure of amyloid fibrils. J Mol Biol 300: 1033–1039. - PubMed
-
- Kodali R, Wetzel R (2007) Polymorphism in the intermediates and products of amyloid assembly. Current Opinion in Structural Biology 17: 48–57. - PubMed
-
- Invernizzi G, Papaleo E, Sabate R, Ventura S (2012) Protein aggregation: mechanisms and functional consequences. Int J Biochem Cell Biol. 44: 1541–1554. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

