INDIGO - INtegrated data warehouse of microbial genomes with examples from the red sea extremophiles
- PMID: 24324765
- PMCID: PMC3855842
- DOI: 10.1371/journal.pone.0082210
INDIGO - INtegrated data warehouse of microbial genomes with examples from the red sea extremophiles
Abstract
Background: The next generation sequencing technologies substantially increased the throughput of microbial genome sequencing. To functionally annotate newly sequenced microbial genomes, a variety of experimental and computational methods are used. Integration of information from different sources is a powerful approach to enhance such annotation. Functional analysis of microbial genomes, necessary for downstream experiments, crucially depends on this annotation but it is hampered by the current lack of suitable information integration and exploration systems for microbial genomes.
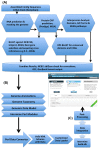
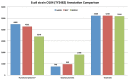
Results: We developed a data warehouse system (INDIGO) that enables the integration of annotations for exploration and analysis of newly sequenced microbial genomes. INDIGO offers an opportunity to construct complex queries and combine annotations from multiple sources starting from genomic sequence to protein domain, gene ontology and pathway levels. This data warehouse is aimed at being populated with information from genomes of pure cultures and uncultured single cells of Red Sea bacteria and Archaea. Currently, INDIGO contains information from Salinisphaera shabanensis, Haloplasma contractile, and Halorhabdus tiamatea - extremophiles isolated from deep-sea anoxic brine lakes of the Red Sea. We provide examples of utilizing the system to gain new insights into specific aspects on the unique lifestyle and adaptations of these organisms to extreme environments.
Conclusions: We developed a data warehouse system, INDIGO, which enables comprehensive integration of information from various resources to be used for annotation, exploration and analysis of microbial genomes. It will be regularly updated and extended with new genomes. It is aimed to serve as a resource dedicated to the Red Sea microbes. In addition, through INDIGO, we provide our Automatic Annotation of Microbial Genomes (AAMG) pipeline. The INDIGO web server is freely available at http://www.cbrc.kaust.edu.sa/indigo.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

