Sex hormone receptor repertoire in breast cancer
- PMID: 24324894
- PMCID: PMC3845405
- DOI: 10.1155/2013/284036
Sex hormone receptor repertoire in breast cancer
Abstract
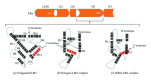
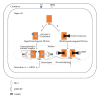
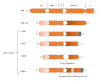
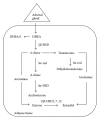
Classification of breast cancer as endocrine sensitive, hormone dependent, or estrogen receptor (ER) positive refers singularly to ER α . One of the oldest recognized tumor targets, disruption of ER α -mediated signaling, is believed to be the mechanistic mode of action for all hormonal interventions used in treating this disease. Whereas ER α is widely accepted as the single most important predictive factor (for response to endocrine therapy), the presence of the receptor in tumor cells is also of prognostic value. Even though the clinical relevance of the two other sex hormone receptors, namely, ER β and the androgen receptor remains unclear, two discordant phenomena observed in hormone-dependent breast cancers could be causally related to ER β -mediated effects and androgenic actions. Nonetheless, our understanding of regulatory molecules and resistance mechanisms remains incomplete, further compromising our ability to develop novel therapeutic strategies that could improve disease outcomes. This review focuses on the receptor-mediated actions of the sex hormones in breast cancer.
Figures

References
-
- Boyd S. On oophorectomy in cancer of the breast. British Medical Journal. 1900;2:1161–1167.
-
- Jensen VE, Jacobson IH, Walf AA, Frye AC. Basic guides to the mechanism of estrogen action. Recent Progress in Hormone Research. 1962;18:318–414.
-
- Fisher B, Redmond C, Fisher ER, Caplan R. Relative worth of estrogen or progesterone receptor and pathologic characteristics of differentiation as indicators of prognosis in node negative breast cancer patients: findings from national surgical adjuvant breast and bowel project protocol B-06. Journal of Clinical Oncology. 1988;6(7):1076–1087. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

