Combination of degradation pathways for naphthalene utilization in Rhodococcus sp. strain TFB
- PMID: 24325207
- PMCID: PMC3937715
- DOI: 10.1111/1751-7915.12096
Combination of degradation pathways for naphthalene utilization in Rhodococcus sp. strain TFB
Abstract
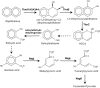
Rhodococcus sp. strain TFB is a metabolic versatile bacterium able to grow on naphthalene as the only carbon and energy source. Applying proteomic, genetic and biochemical approaches, we propose in this paper that, at least, three coordinated but independently regulated set of genes are combined to degrade naphthalene in TFB. First, proteins involved in tetralin degradation are also induced by naphthalene and may carry out its conversion to salicylaldehyde. This is the only part of the naphthalene degradation pathway showing glucose catabolite repression. Second, a salicylaldehyde dehydrogenase activity that converts salicylaldehyde to salicylate is detected in naphthalene-grown cells but not in tetralin- or salicylate-grown cells. Finally, we describe the chromosomally located nag genes, encoding the gentisate pathway for salicylate conversion into fumarate and pyruvate, which are only induced by salicylate and not by naphthalene. This work shows how biodegradation pathways in Rhodococcus sp. strain TFB could be assembled using elements from different pathways mainly because of the laxity of the regulatory systems and the broad specificity of the catabolic enzymes.
© 2013 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Figures




Similar articles
-
Biodegradation of Tetralin: Genomics, Gene Function and Regulation.Genes (Basel). 2019 May 6;10(5):339. doi: 10.3390/genes10050339. Genes (Basel). 2019. PMID: 31064110 Free PMC article. Review.
-
Naphthalene degradation via salicylate and gentisate by Rhodococcus sp. strain B4.Appl Environ Microbiol. 1992 Jun;58(6):1874-7. doi: 10.1128/aem.58.6.1874-1877.1992. Appl Environ Microbiol. 1992. PMID: 1622263 Free PMC article.
-
Molecular and biochemical characterization of the tetralin degradation pathway in Rhodococcus sp. strain TFB.Microb Biotechnol. 2009 Mar;2(2):262-73. doi: 10.1111/j.1751-7915.2009.00086.x. Microb Biotechnol. 2009. PMID: 21261920 Free PMC article.
-
Proteomic and transcriptional characterization of aromatic degradation pathways in Rhodoccocus sp. strain TFB.Proteomics. 2006 Apr;6 Suppl 1:S119-32. doi: 10.1002/pmic.200500422. Proteomics. 2006. PMID: 16544280
-
Biotechnological Potential of Rhodococcus Biodegradative Pathways.J Microbiol Biotechnol. 2018 Jul 28;28(7):1037-1051. doi: 10.4014/jmb.1712.12017. J Microbiol Biotechnol. 2018. PMID: 29913546 Review.
Cited by
-
Rhodococcus Strains from the Specialized Collection of Alkanotrophs for Biodegradation of Aromatic Compounds.Molecules. 2023 Mar 5;28(5):2393. doi: 10.3390/molecules28052393. Molecules. 2023. PMID: 36903638 Free PMC article.
-
Biodegradation of Tetralin: Genomics, Gene Function and Regulation.Genes (Basel). 2019 May 6;10(5):339. doi: 10.3390/genes10050339. Genes (Basel). 2019. PMID: 31064110 Free PMC article. Review.
-
Use of Shotgun Metagenomics to Assess the Microbial Diversity and Hydrocarbons Degrading Functions of Auto-Mechanic Workshops Soils Polluted with Gasoline and Diesel Fuel.Microorganisms. 2023 Mar 10;11(3):722. doi: 10.3390/microorganisms11030722. Microorganisms. 2023. PMID: 36985295 Free PMC article.
-
Time Series Resolution of the Fish Necrobiome Reveals a Decomposer Succession Involving Toxigenic Bacterial Pathogens.mSystems. 2020 Apr 28;5(2):e00145-20. doi: 10.1128/mSystems.00145-20. mSystems. 2020. PMID: 32345738 Free PMC article.
-
Growth modulation and metabolic responses of Ganoderma boninense to salicylic acid stress.PLoS One. 2021 Dec 31;16(12):e0262029. doi: 10.1371/journal.pone.0262029. eCollection 2021. PLoS One. 2021. PMID: 34972183 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

