The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s
- PMID: 24327659
- PMCID: PMC3908422
- DOI: 10.1074/jbc.M113.507491
The large Hsp70 Grp170 binds to unfolded protein substrates in vivo with a regulation distinct from conventional Hsp70s
Abstract
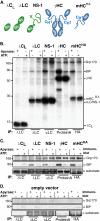
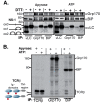
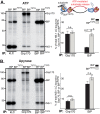
The Hsp70 superfamily is a ubiquitous chaperone class that includes conventional and large Hsp70s. BiP is the only conventional Hsp70 in the endoplasmic reticulum (ER) whose functions include: assisting protein folding, targeting misfolded proteins for degradation, and regulating the transducers of the unfolded protein response. The ER also possesses a single large Hsp70, the glucose-regulated protein of 170 kDa (Grp170). Like BiP it is an essential protein, but its cellular functions are not well understood. Here we show that Grp170 can bind directly to a variety of incompletely folded protein substrates in the ER, and as expected for a bona fide chaperone, it does not interact with folded secretory proteins. Our data demonstrate that Grp170 and BiP associate with similar molecular forms of two substrate proteins, but while BiP is released from unfolded substrates in the presence of ATP, Grp170 remains bound. In comparison to conventional Hsp70s, the large Hsp70s possess two unique structural features: an extended C-terminal α-helical domain and an unstructured loop in the putative substrate binding domain with an unknown function. We find that in the absence of the α-helical domain the interaction of Grp170 with substrates is reduced. In striking contrast, deletion of the unstructured loop results in increased binding to substrates, suggesting the presence of unique intramolecular mechanisms of control for the chaperone functions of large Hsp70s.
Keywords: ER Quality Control; Endoplasmic Reticulum (ER); Glycoprotein; Molecular Cell Biology; Molecular Chaperone; Protein Folding.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

