DeGNServer: deciphering genome-scale gene networks through high performance reverse engineering analysis
- PMID: 24328032
- PMCID: PMC3847961
- DOI: 10.1155/2013/856325
DeGNServer: deciphering genome-scale gene networks through high performance reverse engineering analysis
Abstract
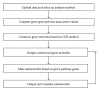
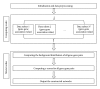
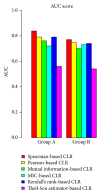
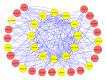
Analysis of genome-scale gene networks (GNs) using large-scale gene expression data provides unprecedented opportunities to uncover gene interactions and regulatory networks involved in various biological processes and developmental programs, leading to accelerated discovery of novel knowledge of various biological processes, pathways and systems. The widely used context likelihood of relatedness (CLR) method based on the mutual information (MI) for scoring the similarity of gene pairs is one of the accurate methods currently available for inferring GNs. However, the MI-based reverse engineering method can achieve satisfactory performance only when sample size exceeds one hundred. This in turn limits their applications for GN construction from expression data set with small sample size. We developed a high performance web server, DeGNServer, to reverse engineering and decipher genome-scale networks. It extended the CLR method by integration of different correlation methods that are suitable for analyzing data sets ranging from moderate to large scale such as expression profiles with tens to hundreds of microarray hybridizations, and implemented all analysis algorithms using parallel computing techniques to infer gene-gene association at extraordinary speed. In addition, we integrated the SNBuilder and GeNa algorithms for subnetwork extraction and functional module discovery. DeGNServer is publicly and freely available online.
Figures
Similar articles
-
Reverse engineering and analysis of genome-wide gene regulatory networks from gene expression profiles using high-performance computing.IEEE/ACM Trans Comput Biol Bioinform. 2012 May-Jun;9(3):668-78. doi: 10.1109/TCBB.2011.60. IEEE/ACM Trans Comput Biol Bioinform. 2012. PMID: 21464509
-
STARNET 2: a web-based tool for accelerating discovery of gene regulatory networks using microarray co-expression data.BMC Bioinformatics. 2009 Oct 14;10:332. doi: 10.1186/1471-2105-10-332. BMC Bioinformatics. 2009. PMID: 19828039 Free PMC article.
-
How to infer gene networks from expression profiles.Mol Syst Biol. 2007;3:78. doi: 10.1038/msb4100120. Epub 2007 Feb 13. Mol Syst Biol. 2007. PMID: 17299415 Free PMC article.
-
Computational biology of genome expression and regulation--a review of microarray bioinformatics.J Environ Pathol Toxicol Oncol. 2008;27(3):157-79. doi: 10.1615/jenvironpatholtoxicoloncol.v27.i3.10. J Environ Pathol Toxicol Oncol. 2008. PMID: 18652564 Review.
-
Inferring regulatory networks.Front Biosci. 2008 Jan 1;13:263-75. doi: 10.2741/2677. Front Biosci. 2008. PMID: 17981545 Review.
Cited by
-
Mining Functional Modules in Heterogeneous Biological Networks Using Multiplex PageRank Approach.Front Plant Sci. 2016 Jun 22;7:903. doi: 10.3389/fpls.2016.00903. eCollection 2016. Front Plant Sci. 2016. PMID: 27446133 Free PMC article.
-
The personal touch: strategies toward personalized vaccines and predicting immune responses to them.Expert Rev Vaccines. 2014 May;13(5):657-69. doi: 10.1586/14760584.2014.905744. Epub 2014 Apr 4. Expert Rev Vaccines. 2014. PMID: 24702429 Free PMC article. Review.
-
LegumeIP 2.0--a platform for the study of gene function and genome evolution in legumes.Nucleic Acids Res. 2016 Jan 4;44(D1):D1189-94. doi: 10.1093/nar/gkv1237. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578557 Free PMC article.
-
Construction and Optimization of a Large Gene Coexpression Network in Maize Using RNA-Seq Data.Plant Physiol. 2017 Sep;175(1):568-583. doi: 10.1104/pp.17.00825. Epub 2017 Aug 2. Plant Physiol. 2017. PMID: 28768814 Free PMC article.
-
Methanol regulated yeast promoters: production vehicles and toolbox for synthetic biology.Microb Cell Fact. 2015 Dec 2;14:196. doi: 10.1186/s12934-015-0387-1. Microb Cell Fact. 2015. PMID: 26627685 Free PMC article.
References
-
- Zhu XL, Ai ZH, Wang J, Xu YL, Teng YC. Weighted gene co-expression network analysis in identification of endometrial cancer prognosis markers. Asian Pacific Journal Cancer Prevention. 2012;13(9):4607–4611. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

