Biosynthesis of nitrogenase metalloclusters
- PMID: 24328215
- PMCID: PMC3999185
- DOI: 10.1021/cr400463x
Biosynthesis of nitrogenase metalloclusters
Conflict of interest statement
The author declares no competing financial interest.
Figures







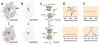
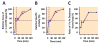
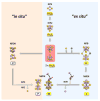
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

