Mechanism of cytotoxic action of crambescidin-816 on human liver-derived tumour cells
- PMID: 24328908
- PMCID: PMC3966746
- DOI: 10.1111/bph.12552
Mechanism of cytotoxic action of crambescidin-816 on human liver-derived tumour cells
Abstract
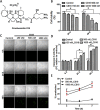
Background and purpose: Marine sponges have evolved the capacity to produce a series of very efficient chemicals to combat viruses, bacteria, and eukaryotic organisms. It has been demonstrated that several of these compounds have anti-neoplastic activity. The highly toxic sponge Crambe crambe has been the source of several molecules named crambescidins. Of these, crambescidin-816 has been shown to be cytotoxic for colon carcinoma cells. To further investigate the potential anti-carcinogenic effect of crambescidin-816, we analysed its effect on the transcription of HepG2 cells by microarray analysis followed by experiments guided by the results obtained.
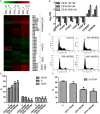
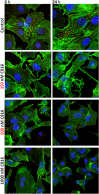
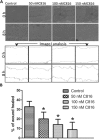
Experimental approach: After cytotoxicity determination, a transcriptomic analysis was performed to test the effect of crambescidin-816 on the liver-derived tumour cell HepG2. Based on the results obtained, we analysed the effect of crambescidin-816 on cell-cell adhesion, cell-matrix adhesion, and cell migration by Western blot, confocal microscopy, flow cytometry and transmission electron microscopy. Cytotoxicity and cell migration were also studied in a variety of other cell lines derived from human tumours.
Key results: Crambescidin-816 had a cytotoxic effect on all the cell lines studied. It inhibited cell-cell adhesion, interfered with the formation of tight junctions, and cell-matrix adhesion, negatively affecting focal adhesions. It also altered the cytoskeleton dynamics. As a consequence of all these effects on cells crambescidin-816 inhibited cell migration.
Conclusions and implications: The results indicate that crambescidin-816 is active against tumour cells and implicate a new mechanism for the anti-tumour effect of this compound.
Keywords: crambescidin-816; transcriptomic analysis; tumour cell adhesion; tumour cell migration; tumour cell viability.
© 2013 The British Pharmacological Society.
Figures





References
-
- Aoki S, Kong D, Matsui K, Kobayashi M. Erythroid differentiation in K562 chronic myelogenous cells induced by crambescidin 800, a pentacyclic guanidine alkaloid. Anticancer Res. 2004;24:2325–2330. - PubMed
-
- Aron ZD, Pietraszkiewicz H, Overman LE, Valeriote F, Cuevas C. Synthesis and anticancer activity of side chain analogs of the crambescidin alkaloids. Bioorg Med Chem Lett. 2004;14:3445–3449. - PubMed
-
- Bates S, Bonetta L, MacAllan D, Parry D, Holder A, Dickson C, et al. CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene. 1994;9:71–79. - PubMed
-
- Belarbi el H, Contreras Gomez A, Chisti Y, Garcia Camacho F, Molina Grima E. Producing drugs from marine sponges. Biotechnol Adv. 2003;21:585–598. - PubMed
-
- Berlinck RG, Braekman JC, Daloze D, Bruno I, Riccio R, Ferri S, et al. Polycyclic guanidine alkaloids from the marine sponge Crambe crambe and Ca++ channel blocker activity of crambescidin 816. J Nat Prod. 1993;56:1007–1015. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

