Chronic ethanol consumption modulates growth factor release, mucosal cytokine production, and microRNA expression in nonhuman primates
- PMID: 24329418
- PMCID: PMC3984381
- DOI: 10.1111/acer.12325
Chronic ethanol consumption modulates growth factor release, mucosal cytokine production, and microRNA expression in nonhuman primates
Abstract
Background: Chronic alcohol consumption has been associated with enhanced susceptibility to both systemic and mucosal infections. However, the exact mechanisms underlying this enhanced susceptibility remain incompletely understood.
Methods: Using a nonhuman primate model of ethanol (EtOH) self-administration, we examined the impact of chronic alcohol exposure on immune homeostasis, cytokine, and growth factor production in peripheral blood, lung, and intestinal mucosa following 12 months of chronic EtOH exposure.
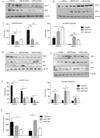
Results: EtOH exposure inhibited activation-induced production of growth factors hepatocyte growth factor (HGF), granulocyte colony-stimulating factor (G-CSF), and vascular-endothelial growth factor (VEGF) by peripheral blood mononuclear cells (PBMC). Moreover, EtOH significantly reduced the frequency of colonic Th1 and Th17 cells in a dose-dependent manner. In contrast, we did not observe differences in lymphocyte frequency or soluble factor production in the lung of EtOH-consuming animals. To uncover mechanisms underlying reduced growth factor and Th1/Th17 cytokine production, we compared expression levels of microRNAs in PBMC and intestinal mucosa. Our analysis revealed EtOH-dependent up-regulation of distinct microRNAs in affected tissues (miR-181a and miR-221 in PBMC; miR-155 in colon). Moreover, we were able to detect reduced expression of the transcription factors STAT3 and ARNT, which regulate expression of VEGF, G-CSF, and HGF and contain targets for these microRNAs. To confirm and extend these observations, PBMC were transfected with either mimics or antagomirs of miR-181 and miR-221, and protein levels of the transcription factors and growth factors were determined. Transfection of microRNA mimics led to a reduction in both STAT3/ARNT as well as VEGF/HGF/G-CSF levels. The opposite outcome was observed when microRNA antagomirs were transfected.
Conclusions: Chronic EtOH consumption significantly disrupts both peripheral and mucosal immune homeostasis, and this dysregulation may be mediated by changes in microRNA expression.
Keywords: Ethanol; Immunity; MicroRNA; Nonhuman Primate; Self-Administration.
Copyright © 2013 by the Research Society on Alcoholism.
Figures






References
-
- Albasanz-Puig A, Murray J, Namekata M, Wijelath ES. Opposing Roles Of Stat-1 And Stat-3 In Regulating Vascular Endothelial Growth Factor Expression In Vascular Smooth Muscle Cells. Biochem Biophys Res Commun. 2012;428:179–184. - PubMed
-
- Bagby G, Zhang P, Stoltz D, Nelson S. Suppression Of The Granulocyte Colony-Stimulating Factor Response To Escherichia Coli Challenge By Alcohol Intoxication. Alcoholism, Clinical And Experimental Research. 1998;22:1740–1745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

