NEMATIC: a simple and versatile tool for the in silico analysis of plant-nematode interactions
- PMID: 24330140
- PMCID: PMC6638708
- DOI: 10.1111/mpp.12114
NEMATIC: a simple and versatile tool for the in silico analysis of plant-nematode interactions
Abstract
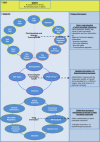
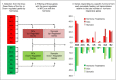
Novel approaches for the control of agriculturally damaging nematodes are sorely needed. Endoparasitic nematodes complete their life cycle within the root vascular cylinder, inducing specialized feeding cells: giant cells for root-knot nematodes and syncytia for cyst nematodes. Both nematodes hijack parts of the transduction cascades involved in developmental processes, or partially mimic the plant responses to other interactions with microorganisms, but molecular evidence of their differences and commonalities is still under investigation. Transcriptomics has been used to describe global expression profiles of their interaction with Arabidopsis, generating vast lists of differentially expressed genes. Although these results are available in public databases and publications, the information is scattered and difficult to handle. Here, we present a rapid, visual, user-friendly and easy to handle spreadsheet tool, called NEMATIC (NEMatode-Arabidopsis Transcriptomic Interaction Compendium; http://www.uclm.es/grupo/gbbmp/english/nematic.asp). It combines existing transcriptomic data for the interaction between Arabidopsis and plant-endoparasitic nematodes with data from different transcriptomic analyses regarding hormone and cell cycle regulation, development, different plant tissues, cell types and various biotic stresses. NEMATIC facilitates efficient in silico studies on plant-nematode biology, allowing rapid cross-comparisons with complex datasets and obtaining customized gene selections through sequential comparative and filtering steps. It includes gene functional classification and links to utilities from several databases. This data-mining spreadsheet will be valuable for the understanding of the molecular bases subjacent to feeding site formation by comparison with other plant systems, and for the selection of genes as potential tools for biotechnological control of nematodes, as demonstrated in the experimentally confirmed examples provided.
Keywords: functional classification; galls; giant cells; in silico analysis; plant-nematode interactions; syncytia; transcriptome comparison.
© 2013 BSPP AND JOHN WILEY & SONS LTD.
Figures
References
-
- Alkharouf, N.W. , Klink, V.P. , Chouikha, I.B. , Beard, H.S. , MacDonald, M.H. , Meyer, S. , Knap, H.T. , Khan, R. and Matthews, B.F. (2006) Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta, 224, 838–852. - PubMed
-
- de Almeida Engler, J. and Favery, B. (2011) The plant cytoskeleton remodelling in nematode induced feeding sites In: Genomics and Molecular Genetics of Plant–Nematode Interactions (Jones J., Gheysen G. and Fenoll C., eds), pp. 369–394. Dordrecht: Springer.
-
- de Almeida Engler, J. , Kyndt, T. , Vieira, P. , Van Cappelle, E. , Boudolf, V. , Sanchez, V. , Escobar, C. , De Veylder, L. , Engler, G. , Abad, P. and Gheysen, G. (2012) CCS52 and DEL1 genes are key components of the endocycle in nematode‐induced feeding sites. Plant J. 72, 185–198. - PubMed
-
- Barcala, M. , Garcia, A. , Cabrera, J. , Casson, S. , Lindsey, K. , Favery, B. , García‐Casado, G. , Solano, R. , Fenoll, C. and Escobar, C. (2010) Early transcriptomic events in microdissected Arabidopsis nematode‐induced giant cells. Plant J. 61, 698–712. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

