Genome-wide mapping of DNA methylation in the human malaria parasite Plasmodium falciparum
- PMID: 24331467
- PMCID: PMC3931529
- DOI: 10.1016/j.chom.2013.11.007
Genome-wide mapping of DNA methylation in the human malaria parasite Plasmodium falciparum
Abstract
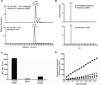
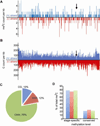
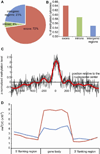
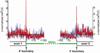
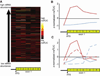
Cytosine DNA methylation is an epigenetic mark in most eukaryotic cells that regulates numerous processes, including gene expression and stress responses. We performed a genome-wide analysis of DNA methylation in the human malaria parasite Plasmodium falciparum. We mapped the positions of methylated cytosines and identified a single functional DNA methyltransferase (Plasmodium falciparum DNA methyltransferase; PfDNMT) that may mediate these genomic modifications. These analyses revealed that the malaria genome is asymmetrically methylated and shares common features with undifferentiated plant and mammalian cells. Notably, core promoters are hypomethylated, and transcript levels correlate with intraexonic methylation. Additionally, there are sharp methylation transitions at nucleosome and exon-intron boundaries. These data suggest that DNA methylation could regulate virulence gene expression and transcription elongation. Furthermore, the broad range of action of DNA methylation and the uniqueness of PfDNMT suggest that the methylation pathway is a potential target for antimalarial strategies.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Discovery of a new predominant cytosine DNA modification that is linked to gene expression in malaria parasites.Nucleic Acids Res. 2020 Jan 10;48(1):184-199. doi: 10.1093/nar/gkz1093. Nucleic Acids Res. 2020. PMID: 31777939 Free PMC article.
-
Proteomic Identification and Analysis of Arginine-Methylated Proteins of Plasmodium falciparum at Asexual Blood Stages.J Proteome Res. 2017 Feb 3;16(2):368-383. doi: 10.1021/acs.jproteome.5b01052. Epub 2017 Jan 3. J Proteome Res. 2017. PMID: 27933903
-
The Architectural Factor HMGB1 Is Involved in Genome Organization in the Human Malaria Parasite Plasmodium falciparum.mBio. 2021 Apr 27;12(2):e00148-21. doi: 10.1128/mBio.00148-21. mBio. 2021. PMID: 33906919 Free PMC article.
-
Epigenetic Players of Chromatin Structure Regulation in Plasmodium falciparum.Chembiochem. 2019 May 15;20(10):1225-1230. doi: 10.1002/cbic.201800718. Epub 2019 Mar 4. Chembiochem. 2019. PMID: 30632244 Review.
-
Curation of the Plasmodium falciparum genome.Trends Parasitol. 2004 Dec;20(12):548-52. doi: 10.1016/j.pt.2004.09.003. Trends Parasitol. 2004. PMID: 15522662 Review.
Cited by
-
Adenine DNA methylation, 3D genome organization, and gene expression in the parasite Trichomonas vaginalis.Proc Natl Acad Sci U S A. 2020 Jun 9;117(23):13033-13043. doi: 10.1073/pnas.1917286117. Epub 2020 May 27. Proc Natl Acad Sci U S A. 2020. PMID: 32461362 Free PMC article.
-
Rapid activation of distinct members of multigene families in Plasmodium spp.Commun Biol. 2020 Jul 3;3(1):351. doi: 10.1038/s42003-020-1081-3. Commun Biol. 2020. PMID: 32620892 Free PMC article.
-
Malaria in the 'Omics Era'.Genes (Basel). 2021 May 30;12(6):843. doi: 10.3390/genes12060843. Genes (Basel). 2021. PMID: 34070769 Free PMC article. Review.
-
Transcriptomic changes in the microsporidia proliferation and host responses in congenitally infected embryos and larvae.BMC Genomics. 2024 Apr 1;25(1):321. doi: 10.1186/s12864-024-10236-y. BMC Genomics. 2024. PMID: 38556880 Free PMC article.
-
Widespread 5-methylcytosine in the genomes of avian Coccidia and other apicomplexan parasites detected by an ELISA-based method.Parasitol Res. 2017 May;116(5):1573-1579. doi: 10.1007/s00436-017-5434-x. Epub 2017 Mar 30. Parasitol Res. 2017. PMID: 28361273
References
-
- Boyko A, Kovalchuk I. Epigenetic control of plant stress response. Environ Mol Mutagen. 2008;49:61–72. - PubMed
-
- Brueckner B, Garcia Boy R, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. - PubMed
-
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

