Exploring the role of receptor flexibility in structure-based drug discovery
- PMID: 24332165
- PMCID: PMC4459653
- DOI: 10.1016/j.bpc.2013.10.007
Exploring the role of receptor flexibility in structure-based drug discovery
Abstract
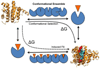
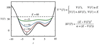
The proper understanding of biomolecular recognition mechanisms that take place in a drug target is of paramount importance to improve the efficiency of drug discovery and development. The intrinsic dynamic character of proteins has a strong influence on biomolecular recognition mechanisms and models such as conformational selection have been widely used to account for this dynamic association process. However, conformational changes occurring in the receptor prior and upon association with other molecules are diverse and not obvious to predict when only a few structures of the receptor are available. In view of the prominent role of protein flexibility in ligand binding and its implications for drug discovery, it is of great interest to identify receptor conformations that play a major role in biomolecular recognition before starting rational drug design efforts. In this review, we discuss a number of recent advances in computer-aided drug discovery techniques that have been proposed to incorporate receptor flexibility into structure-based drug design. The allowance for receptor flexibility provided by computational techniques such as molecular dynamics simulations or enhanced sampling techniques helps to improve the accuracy of methods used to estimate binding affinities and, thus, such methods can contribute to the discovery of novel drug leads.
Keywords: Accelerated molecular dynamics; Allostery; Computer-aided drug design; Conformational selection; Molecular dynamics; Receptor flexibility.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures

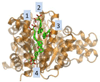
References
-
- Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. Journal of Molecular Biology. 1965;12:88–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

