Collective invasion in breast cancer requires a conserved basal epithelial program
- PMID: 24332913
- PMCID: PMC3941206
- DOI: 10.1016/j.cell.2013.11.029
Collective invasion in breast cancer requires a conserved basal epithelial program
Abstract
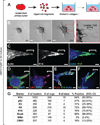
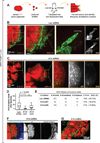
Carcinomas typically invade as a cohesive multicellular unit, a process termed collective invasion. It remains unclear how different subpopulations of cancer cells contribute to this process. We developed three-dimensional (3D) organoid assays to identify the most invasive cancer cells in primary breast tumors. Collective invasion was led by specialized cancer cells that were defined by their expression of basal epithelial genes, such as cytokeratin-14 (K14) and p63. Furthermore, K14+ cells led collective invasion in the major human breast cancer subtypes. Importantly, luminal cancer cells were observed to convert phenotypically to invasive leaders following induction of basal epithelial genes. Although only a minority of cells within luminal tumors expressed basal epithelial genes, knockdown of either K14 or p63 was sufficient to block collective invasion. Our data reveal that heterotypic interactions between epithelial subpopulations are critical to collective invasion. We suggest that targeting the basal invasive program could limit metastatic progression.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





References
-
- Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. 2013;8:277–302. - PubMed
-
- Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. - PubMed
-
- de Silva Rudland S, Platt-Higgins A, Winstanley JH, Jones NJ, Barraclough R, West C, Carroll J, Rudland PS. Statistical association of basal cell keratins with metastasis-inducing proteins in a prognostically unfavorable group of sporadic breast cancers. Am. J. Pathol. 2011;179:1061–1072. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

