A comprehensive analysis of the human placenta transcriptome
- PMID: 24333048
- PMCID: PMC3978120
- DOI: 10.1016/j.placenta.2013.11.007
A comprehensive analysis of the human placenta transcriptome
Abstract
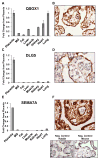
As the conduit for nutrients and growth signals, the placenta is critical to establishing an environment sufficient for fetal growth and development. To better understand the mechanisms regulating placental development and gene expression, we characterized the transcriptome of term placenta from 20 healthy women with uncomplicated pregnancies using RNA-seq. To identify genes that were highly expressed and unique to the placenta we compared placental RNA-seq data to data from 7 other tissues (adipose, breast, hear, kidney, liver, lung, and smooth muscle) and identified several genes novel to placental biology (QSOX1, DLG5, and SEMA7A). Semi-quantitative RT-PCR confirmed the RNA-seq results and immunohistochemistry indicated these proteins were highly expressed in the placental syncytium. Additionally, we mined our RNA-seq data to map the relative expression of key developmental gene families (Fox, Sox, Gata, Tead, and Wnt) within the placenta. We identified FOXO4, GATA3, and WNT7A to be amongst the highest expressed members of these families. Overall, these findings provide a new reference for understanding of placental transcriptome and can aid in the identification of novel pathways regulating placenta physiology that may be dysregulated in placental disease.
Keywords: Development; Fetal tissue; Gene expression; Pregnancy; RNA-seq.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures


References
-
- Sitras V, Paulssen RH, Gronaas H, Vartun A, Acharya G. Gene expression profile in labouring and non-labouring human placenta near term. Mol Hum Reprod. 2008;14(1):61–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

