Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function
- PMID: 24335084
- PMCID: PMC3950677
- DOI: 10.1093/nar/gkt1276
Structure and sequence elements of the CR4/5 domain of medaka telomerase RNA important for telomerase function
Abstract
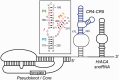
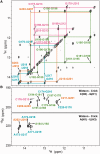
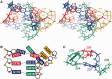
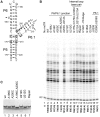
Telomerase is a unique reverse transcriptase that maintains the 3' ends of eukaryotic chromosomes by adding tandem telomeric repeats. The RNA subunit (TR) of vertebrate telomerase provides a template for reverse transcription, contained within the conserved template/pseudoknot domain, and a conserved regions 4 and 5 (CR4/5) domain, all essential for catalytic activity. We report the nuclear magnetic resonance (NMR) solution structure of the full-length CR4/5 domain from the teleost fish medaka (Oryzias latipes). Three helices emanate from a structured internal loop, forming a Y-shaped structure, where helix P6 stacks on P5 and helix P6.1 points away from P6. The relative orientations of the three helices are Mg2+ dependent and dynamic. Although the three-way junction is structured and has unexpected base pairs, telomerase activity assays with nucleotide substitutions and deletions in CR4/5 indicate that none of these are essential for activity. The results suggest that the junction is likely to change conformation in complex with telomerase reverse transcriptase and that it provides a flexible scaffold that allows P6 and P6.1 to correctly fold and interact with telomerase reverse transcriptase.
Figures


Similar articles
-
Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human.Proc Natl Acad Sci U S A. 2016 Aug 30;113(35):E5125-34. doi: 10.1073/pnas.1607411113. Epub 2016 Aug 16. Proc Natl Acad Sci U S A. 2016. PMID: 27531956 Free PMC article.
-
Structural basis for protein-RNA recognition in telomerase.Nat Struct Mol Biol. 2014 Jun;21(6):507-12. doi: 10.1038/nsmb.2819. Epub 2014 May 4. Nat Struct Mol Biol. 2014. PMID: 24793650 Free PMC article.
-
A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA.Nucleic Acids Res. 2002 Jan 15;30(2):592-7. doi: 10.1093/nar/30.2.592. Nucleic Acids Res. 2002. PMID: 11788723 Free PMC article.
-
Structure and function of telomerase RNA.Curr Opin Struct Biol. 2006 Jun;16(3):307-18. doi: 10.1016/j.sbi.2006.05.005. Epub 2006 May 18. Curr Opin Struct Biol. 2006. PMID: 16713250 Review.
-
Telomerase is an unusual RNA-containing enzyme. A review.Biochemistry (Mosc). 1997 Nov;62(11):1206-15. Biochemistry (Mosc). 1997. PMID: 9467844 Review.
Cited by
-
Functional Interactions of Kluyveromyces lactis Telomerase Reverse Transcriptase with the Three-Way Junction and the Template Domains of Telomerase RNA.Int J Mol Sci. 2022 Sep 15;23(18):10757. doi: 10.3390/ijms231810757. Int J Mol Sci. 2022. PMID: 36142669 Free PMC article.
-
Single-molecule FRET-Rosetta reveals RNA structural rearrangements during human telomerase catalysis.RNA. 2017 Feb;23(2):175-188. doi: 10.1261/rna.058743.116. Epub 2016 Nov 15. RNA. 2017. PMID: 28096444 Free PMC article.
-
Influence of Na+ and Mg2+ ions on RNA structures studied with molecular dynamics simulations.Nucleic Acids Res. 2018 Jun 1;46(10):4872-4882. doi: 10.1093/nar/gky221. Nucleic Acids Res. 2018. PMID: 29718375 Free PMC article.
-
Structural Roles of Noncoding RNAs in the Heart of Enzymatic Complexes.Biochemistry. 2017 Jan 10;56(1):3-13. doi: 10.1021/acs.biochem.6b01106. Epub 2016 Dec 29. Biochemistry. 2017. PMID: 27935277 Free PMC article.
-
Structural biology of telomeres and telomerase.Cell Mol Life Sci. 2020 Jan;77(1):61-79. doi: 10.1007/s00018-019-03369-x. Epub 2019 Nov 14. Cell Mol Life Sci. 2020. PMID: 31728577 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

