SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage
- PMID: 24335234
- PMCID: PMC3924925
- DOI: 10.1182/blood-2013-04-490847
SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage
Abstract
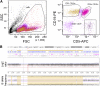
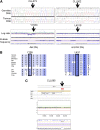
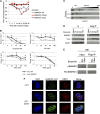
SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase and a nuclease that restricts HIV-1 in noncycling cells. Germ-line mutations in SAMHD1 have been described in patients with Aicardi-Goutières syndrome (AGS), a congenital autoimmune disease. In a previous longitudinal whole genome sequencing study of chronic lymphocytic leukemia (CLL), we revealed a SAMHD1 mutation as a potential founding event. Here, we describe an AGS patient carrying a pathogenic germ-line SAMHD1 mutation who developed CLL at 24 years of age. Using clinical trial samples, we show that acquired SAMHD1 mutations are associated with high variant allele frequency and reduced SAMHD1 expression and occur in 11% of relapsed/refractory CLL patients. We provide evidence that SAMHD1 regulates cell proliferation and survival and engages in specific protein interactions in response to DNA damage. We propose that SAMHD1 may have a function in DNA repair and that the presence of SAMHD1 mutations in CLL promotes leukemia development.
Figures


Comment in
-
SAMHD1: a new gene for CLL.Blood. 2014 Feb 13;123(7):951-2. doi: 10.1182/blood-2013-12-545384. Blood. 2014. PMID: 24526775 No abstract available.
References
-
- Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. - PubMed
-
- Quesada V, Conde L, Villamor N, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2012;44(1):47–52. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

