Molecular evolution of protein-RNA mimicry as a mechanism for translational control
- PMID: 24335280
- PMCID: PMC3950694
- DOI: 10.1093/nar/gkt1296
Molecular evolution of protein-RNA mimicry as a mechanism for translational control
Abstract
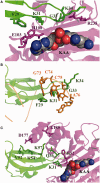
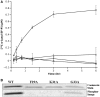
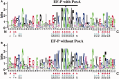
Elongation factor P (EF-P) is a conserved ribosome-binding protein that structurally mimics tRNA to enable the synthesis of peptides containing motifs that otherwise would induce translational stalling, including polyproline. In many bacteria, EF-P function requires post-translational modification with (R)-β-lysine by the lysyl-tRNA synthetase paralog PoxA. To investigate how recognition of EF-P by PoxA evolved from tRNA recognition by aminoacyl-tRNA synthetases, we compared the roles of EF-P/PoxA polar contacts with analogous interactions in a closely related tRNA/synthetase complex. PoxA was found to recognize EF-P solely via identity elements in the acceptor loop, the domain of the protein that interacts with the ribosome peptidyl transferase center and mimics the 3'-acceptor stem of tRNA. Although the EF-P acceptor loop residues required for PoxA recognition are highly conserved, their conservation was found to be independent of the phylogenetic distribution of PoxA. This suggests EF-P first evolved tRNA mimicry to optimize interactions with the ribosome, with PoxA-catalyzed aminoacylation evolving later as a secondary mechanism to further improve ribosome binding and translation control.
Figures
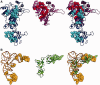


Similar articles
-
The tRNA synthetase paralog PoxA modifies elongation factor-P with (R)-β-lysine.Nat Chem Biol. 2011 Aug 14;7(10):667-9. doi: 10.1038/nchembio.632. Nat Chem Biol. 2011. PMID: 21841797 Free PMC article.
-
A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P.Nat Struct Mol Biol. 2010 Sep;17(9):1136-43. doi: 10.1038/nsmb.1889. Epub 2010 Aug 22. Nat Struct Mol Biol. 2010. PMID: 20729861
-
Structural Basis for Polyproline-Mediated Ribosome Stalling and Rescue by the Translation Elongation Factor EF-P.Mol Cell. 2017 Nov 2;68(3):515-527.e6. doi: 10.1016/j.molcel.2017.10.014. Mol Cell. 2017. PMID: 29100052
-
[Elongation Factor P: New Mechanisms of Function and an Evolutionary Diversity of Translation Regulation].Mol Biol (Mosk). 2019 Jul-Aug;53(4):561-573. doi: 10.1134/S0026898419040037. Mol Biol (Mosk). 2019. PMID: 31397432 Review. Russian.
-
Is tRNA binding or tRNA mimicry mandatory for translation factors?Curr Protein Pept Sci. 2002 Feb;3(1):133-41. doi: 10.2174/1389203023380837. Curr Protein Pept Sci. 2002. PMID: 12370017 Review.
Cited by
-
Carbonyl reduction by YmfI in Bacillus subtilis prevents accumulation of an inhibitory EF-P modification state.Mol Microbiol. 2017 Oct;106(2):236-251. doi: 10.1111/mmi.13760. Epub 2017 Aug 22. Mol Microbiol. 2017. PMID: 28787546 Free PMC article.
-
The non-canonical hydroxylase structure of YfcM reveals a metal ion-coordination motif required for EF-P hydroxylation.Nucleic Acids Res. 2014 Oct 29;42(19):12295-305. doi: 10.1093/nar/gku898. Epub 2014 Oct 1. Nucleic Acids Res. 2014. PMID: 25274739 Free PMC article.
-
Structural Basis for EarP-Mediated Arginine Glycosylation of Translation Elongation Factor EF-P.mBio. 2017 Sep 26;8(5):e01412-17. doi: 10.1128/mBio.01412-17. mBio. 2017. PMID: 28951478 Free PMC article.
-
Neisseria meningitidis Translation Elongation Factor P and Its Active-Site Arginine Residue Are Essential for Cell Viability.PLoS One. 2016 Feb 3;11(2):e0147907. doi: 10.1371/journal.pone.0147907. eCollection 2016. PLoS One. 2016. PMID: 26840407 Free PMC article.
-
Quality Control Mechanisms in Bacterial Translation.Acta Naturae. 2021 Apr-Jun;13(2):32-44. doi: 10.32607/actanaturae.11401. Acta Naturae. 2021. PMID: 34377554 Free PMC article.
References
-
- Pasteur G. A classificatory review of mimicry systems. Annu. Rev. Ecol. Syst. 1982;13:169–199.
-
- Elde NC, Malik HS. The evolutionary conundrum of pathogen mimicry. Nat. Rev. Microbiol. 2009;7:787–797. - PubMed
-
- Keene JD. RNA surfaces as functional mimetics of proteins. Chem. Biol. 1996;3:505–513. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

