Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication
- PMID: 24335281
- PMCID: PMC3950690
- DOI: 10.1093/nar/gkt1292
Genetic analysis of the Replication Protein A large subunit family in Arabidopsis reveals unique and overlapping roles in DNA repair, meiosis and DNA replication
Abstract
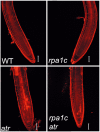
Replication Protein A (RPA) is a heterotrimeric protein complex that binds single-stranded DNA. In plants, multiple genes encode the three RPA subunits (RPA1, RPA2 and RPA3), including five RPA1-like genes in Arabidopsis. Phylogenetic analysis suggests two distinct groups composed of RPA1A, RPA1C, RPA1E (ACE group) and RPA1B, RPA1D (BD group). ACE-group members are transcriptionally induced by ionizing radiation, while BD-group members show higher basal transcription and are not induced by ionizing radiation. Analysis of rpa1 T-DNA insertion mutants demonstrates that although each mutant line is likely null, all mutant lines are viable and display normal vegetative growth. The rpa1c and rpa1e single mutants however display hypersensitivity to ionizing radiation, and combination of rpa1c and rpa1e results in additive hypersensitivity to a variety of DNA damaging agents. Combination of the partially sterile rpa1a with rpa1c results in complete sterility, incomplete synapsis and meiotic chromosome fragmentation, suggesting an early role for RPA1C in promoting homologous recombination. Combination of either rpa1c and/or rpa1e with atr revealed additive hypersensitivity phenotypes consistent with each functioning in unique repair pathways. In contrast, rpa1b rpa1d double mutant plants display slow growth and developmental defects under non-damaging conditions. We show these defects in the rpa1b rpa1d mutant are likely the result of defective DNA replication leading to reduction in cell division.
Figures






References
-
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Ann. Rev. Biochem. 1997;66:61–92. - PubMed
-
- Abraham R. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. - PubMed
-
- Hurley PJ, Bunz F. ATM and ATR: components of an integrated circuit. Cell cycle (Georgetown, Tex) 2007;6:414–417. - PubMed
-
- Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 2002;14:237–245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

