Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication
- PMID: 24335297
- PMCID: PMC3911523
- DOI: 10.1128/JVI.03443-13
Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication
Abstract
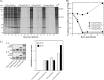
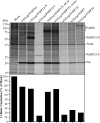
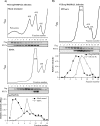
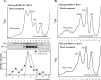
The innate immune response is the first line of defense against most viral infections. Its activation promotes cell signaling, which reduces virus replication in infected cells and leads to induction of the antiviral state in yet-uninfected cells. This inhibition of virus replication is a result of the activation of a very broad spectrum of specific cellular genes, with each of their products usually making a small but detectable contribution to the overall antiviral state. The lack of a strong, dominant function for each gene product and the ability of many viruses to interfere with the development of the antiviral response strongly complicate identification of the antiviral activity of the activated individual cellular genes. However, we have previously developed and applied a new experimental system which allows us to define a critical function of some members of the poly(ADP-ribose) polymerase (PARP) family in clearance of Venezuelan equine encephalitis virus mutants from infected cells. In this new study, we demonstrate that PARP7, PARP10, and the long isoform of PARP12 (PARP12L) function as important and very potent regulators of cellular translation and virus replication. The translation inhibition and antiviral effect of PARP12L appear to be mediated by more than one protein function and are a result of its direct binding to polysomes, complex formation with cellular RNAs (which is determined by both putative RNA-binding and PARP domains), and catalytic activity. IMPORTANCE The results of this study demonstrate that interferon-stimulated gene products PARP7, PARP10, and PARP12L are potent inhibitors of the replication of Venezuelan equine encephalitis virus and other alphaviruses. The inhibitory functions are determined by more than a single mechanism, and one of them is based on the ability of these proteins to regulate cellular translation. Interference with the cellular translational machinery depends on the integrity of both the amino-terminal domain, containing a number of putative RNA-binding motifs, and the catalytic function of the carboxy-terminal PARP domain. The PARP-induced changes in translation efficiency appear to have a more potent effect on the synthesis of virus-specific proteins than on that of cellular proteins, thus making PARP-specific translational downregulation an important contributor to the overall development of the antiviral response.
Figures




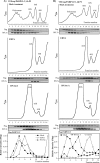
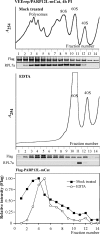
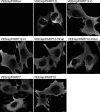
References
-
- Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O'Guin AK, Schmidt RE, Levine B, Virgin HW., IV 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974–13983. 10.1128/JVI.79.22.13974-13983.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

