Enhanced fusion and virion incorporation for HIV-1 subtype C envelope glycoproteins with compact V1/V2 domains
- PMID: 24335304
- PMCID: PMC3911571
- DOI: 10.1128/JVI.02308-13
Enhanced fusion and virion incorporation for HIV-1 subtype C envelope glycoproteins with compact V1/V2 domains
Abstract
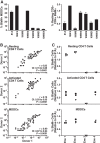
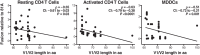
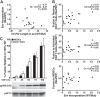
In infected people, the HIV-1 envelope glycoprotein (Env) constantly evolves to escape the immune response while retaining the essential elements needed to mediate viral entry into target cells. The extensive genetic variation of Env is particularly striking in the V1/V2 hypervariable domains. In this study, we investigated the trade-off, in terms of fusion efficiency, for encoding V1/V2 domains of different lengths. We found that natural variations in V1/V2 length exert a profound impact on HIV-1 entry. Variants encoding compact V1/V2 domains mediated fusion with higher efficiencies than related Envs encoding longer V1/V2 domains. By exchanging the V1/V2 domains between Envs of the same infected person or between two persons linked by a transmission event, we further demonstrated that V1/V2 domains critically influence both Env incorporation into viral particles and fusion to primary CD4 T cells and monocyte-derived dendritic cells. Shortening the V1/V2 domains consistently increased Env incorporation and fusion, whereas lengthening the V1/V2 domains decreased Env incorporation and fusion. Given that in a new host transmitted founder viruses are distinguished by compact Envs with fewer glycosylation sites, our study points to fusion and possibly Env incorporation into virions as limiting steps for transmission of HIV-1 to a new host and suggests that the length and/or the N-glycosylation profile of the V1/V2 domain influences these early steps in the HIV life cycle.
Figures





Similar articles
-
A model for alignment of Env V1 and V2 hypervariable domains from human and simian immunodeficiency viruses.AIDS Res Hum Retroviruses. 1996 Aug 10;12(12):1169-78. doi: 10.1089/aid.1996.12.1169. AIDS Res Hum Retroviruses. 1996. PMID: 8844021
-
Turnover of env variable region 1 and 2 genotypes in subjects with late-stage human immunodeficiency virus type 1 infection.J Virol. 2003 Jun;77(12):6811-22. doi: 10.1128/jvi.77.12.6811-6822.2003. J Virol. 2003. PMID: 12768001 Free PMC article.
-
Independent variation and positive selection in env V1 and V2 domains within maternal-infant strains of human immunodeficiency virus type 1 in vivo.J Virol. 1993 Jul;67(7):3951-60. doi: 10.1128/JVI.67.7.3951-3960.1993. J Virol. 1993. PMID: 8510212 Free PMC article.
-
Comparative analysis of the fusion efficiency elicited by the envelope glycoprotein V1-V5 regions derived from human immunodeficiency virus type 1 transmitted perinatally.J Gen Virol. 2012 Dec;93(Pt 12):2635-2645. doi: 10.1099/vir.0.046771-0. Epub 2012 Sep 5. J Gen Virol. 2012. PMID: 22956734 Free PMC article.
-
Diversification in the HIV-1 Envelope Hyper-variable Domains V2, V4, and V5 and Higher Probability of Transmitted/Founder Envelope Glycosylation Favor the Development of Heterologous Neutralization Breadth.PLoS Pathog. 2016 Nov 16;12(11):e1005989. doi: 10.1371/journal.ppat.1005989. eCollection 2016 Nov. PLoS Pathog. 2016. PMID: 27851829 Free PMC article.
Cited by
-
The evolution of envelope function during coinfection with phylogenetically distinct human immunodeficiency virus.BMC Infect Dis. 2024 Sep 9;24(1):934. doi: 10.1186/s12879-024-09805-z. BMC Infect Dis. 2024. PMID: 39251948 Free PMC article.
-
In silico Analysis of HIV-1 Env-gp120 Reveals Structural Bases for Viral Adaptation in Growth-Restrictive Cells.Front Microbiol. 2016 Feb 9;7:110. doi: 10.3389/fmicb.2016.00110. eCollection 2016. Front Microbiol. 2016. PMID: 26903989 Free PMC article.
-
Molecular Features of the V1-V4 Coding Region of Sexually Transmitted Human Immunodeficiency Virus Type 1.J Infect Dis. 2017 May 15;215(10):1506-1513. doi: 10.1093/infdis/jix184. J Infect Dis. 2017. PMID: 28419276 Free PMC article.
-
The HIV-1 transmission bottleneck.Retrovirology. 2017 Mar 23;14(1):22. doi: 10.1186/s12977-017-0343-8. Retrovirology. 2017. PMID: 28335782 Free PMC article.
-
HIV-1 Fusion Assay.Bio Protoc. 2014 Aug 20;4(16):e1212. doi: 10.21769/bioprotoc.1212. Bio Protoc. 2014. PMID: 27525294 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

