Highly divergent integration profile of adeno-associated virus serotype 5 revealed by high-throughput sequencing
- PMID: 24335317
- PMCID: PMC3958079
- DOI: 10.1128/JVI.03419-13
Highly divergent integration profile of adeno-associated virus serotype 5 revealed by high-throughput sequencing
Abstract
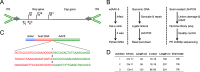
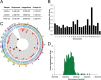
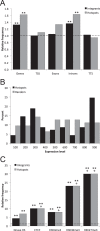
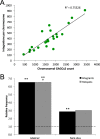
Adeno-associated virus serotype 5 (AAV-5) is a human parvovirus that infects a high percentage of the population. It is the most divergent AAV, the DNA sequence cleaved by the viral endonuclease is distinct from all other described serotypes and, uniquely, AAV-5 does not cross-complement the replication of other serotypes. In contrast to the well-characterized integration of AAV-2, no published studies have investigated the genomic integration of AAV-5. In this study, we analyzed more than 660,000 AAV-5 integration junctions using high-throughput integrant capture sequencing of infected human cells. The integration activity of AAV-5 was 99.7% distinct from AAV-2 and favored intronic sequences. Genome-wide integration was highly correlated with viral replication protein binding and endonuclease sites, and a 39-bp consensus integration motif was revealed that included these features. Algorithmic scanning identified 126 AAV-5 hot spots, the largest of which encompassed 3.3% of all integration events. The unique aspects of AAV-5 integration may provide novel tools for biotechnology and gene therapy.
Importance: Viral integration into the host genome is an important aspect of virus host cell biology. Genomic integration studies of the small single-stranded AAVs have largely focused on site preferential integration of AAV-2, which depends on the viral replication protein (Rep). We have now established the first genome wide integration profile of the highly divergent AAV-5 serotype. Using integrant capture sequencing, more than 600,000 AAV-5 integration junctions in human cells were analyzed. AAV-5 integration hot spots were 99.7% distinct from AAV-2. Integration favored intronic sequences, occurred on all chromosomes, and integration hot spot distribution was correlated with human genomic GAGC repeats and transcriptional activity. These features support expansion of AAV-5 based vectors for gene transfer considerations.
Figures
Similar articles
-
High-throughput sequencing reveals principles of adeno-associated virus serotype 2 integration.J Virol. 2013 Aug;87(15):8559-68. doi: 10.1128/JVI.01135-13. Epub 2013 May 29. J Virol. 2013. PMID: 23720718 Free PMC article.
-
Adeno-associated virus type 2 wild-type and vector-mediated genomic integration profiles of human diploid fibroblasts analyzed by third-generation PacBio DNA sequencing.J Virol. 2014 Oct;88(19):11253-63. doi: 10.1128/JVI.01356-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031342 Free PMC article.
-
Integration preferences of wildtype AAV-2 for consensus rep-binding sites at numerous loci in the human genome.PLoS Pathog. 2010 Jul 8;6(7):e1000985. doi: 10.1371/journal.ppat.1000985. PLoS Pathog. 2010. PMID: 20628575 Free PMC article.
-
Integration of adeno-associated virus (AAV) and recombinant AAV vectors.Annu Rev Genet. 2004;38:819-45. doi: 10.1146/annurev.genet.37.110801.143717. Annu Rev Genet. 2004. PMID: 15568995 Review.
-
Adeno-associated virus integration: virus versus vector.Gene Ther. 2008 Jun;15(11):817-22. doi: 10.1038/gt.2008.55. Epub 2008 Apr 10. Gene Ther. 2008. PMID: 18401436 Review.
Cited by
-
Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus.PLoS One. 2018 Jul 17;13(7):e0200841. doi: 10.1371/journal.pone.0200841. eCollection 2018. PLoS One. 2018. PMID: 30016371 Free PMC article.
-
Use of Adeno-Associated Virus to Enrich Cardiomyocytes Derived from Human Stem Cells.Hum Gene Ther Clin Dev. 2015 Sep;26(3):194-201. doi: 10.1089/humc.2015.052. Epub 2015 Aug 7. Hum Gene Ther Clin Dev. 2015. PMID: 26252064 Free PMC article.
-
ITR-Seq, a next-generation sequencing assay, identifies genome-wide DNA editing sites in vivo following adeno-associated viral vector-mediated genome editing.BMC Genomics. 2020 Mar 17;21(1):239. doi: 10.1186/s12864-020-6655-4. BMC Genomics. 2020. PMID: 32183699 Free PMC article.
-
Parvovirus B19 integration into human CD36+ erythroid progenitor cells.Virology. 2017 Nov;511:40-48. doi: 10.1016/j.virol.2017.08.011. Epub 2017 Aug 12. Virology. 2017. PMID: 28806616 Free PMC article.
-
Enhort: a platform for deep analysis of genomic positions.PeerJ Comput Sci. 2019 Jun 10;5:e198. doi: 10.7717/peerj-cs.198. eCollection 2019. PeerJ Comput Sci. 2019. PMID: 33816851 Free PMC article.
References
-
- Halbert CL, Miller AD, McNamara S, Emerson J, Gibson RL, Ramsey B, Aitken ML. 2006. Prevalence of neutralizing antibodies against adeno-associated virus (AAV) types 2, 5, and 6 in cystic fibrosis and normal populations: implications for gene therapy using AAV vectors. Hum. Gene Ther. 17:440–447. 10.1089/hum.2006.17.440 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

