A conserved KIN17 curved DNA-binding domain protein assembles with SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 to adapt Arabidopsis growth and development to limiting copper availability
- PMID: 24335506
- PMCID: PMC3912109
- DOI: 10.1104/pp.113.228239
A conserved KIN17 curved DNA-binding domain protein assembles with SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 to adapt Arabidopsis growth and development to limiting copper availability
Abstract
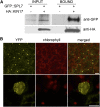
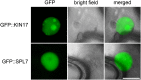
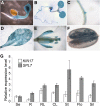
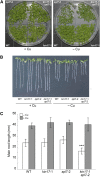
Proper copper (Cu) homeostasis is required by living organisms to maintain essential cellular functions. In the model plant Arabidopsis (Arabidopsis thaliana), the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE7 (SPL7) transcription factor participates in reprogramming global gene expression during Cu insufficiency in order to improve the metal uptake and prioritize its distribution to Cu proteins of major importance. As a consequence, spl7 null mutants show morphological and physiological disorders during Cu-limited growth, resulting in lower fresh weight, reduced root elongation, and chlorosis. On the other hand, the Arabidopsis KIN17 homolog belongs to a well-conserved family of essential eukaryotic nuclear proteins known to be stress activated and involved in DNA and possibly RNA metabolism in mammals. In the study presented here, we uncovered that Arabidopsis KIN17 participates in promoting the Cu deficiency response by means of a direct interaction with SPL7. Moreover, the double mutant kin17-1 spl7-2 displays an enhanced Cu-dependent phenotype involving growth arrest, oxidative stress, floral bud abortion, and pollen inviability. Taken together, the data presented here provide evidence for SPL7 and KIN17 protein interaction as a point of convergence in response to both Cu deficiency and oxidative stress.
Figures





Similar articles
-
Arabidopsis Pollen Fertility Requires the Transcription Factors CITF1 and SPL7 That Regulate Copper Delivery to Anthers and Jasmonic Acid Synthesis.Plant Cell. 2017 Dec;29(12):3012-3029. doi: 10.1105/tpc.17.00363. Epub 2017 Nov 7. Plant Cell. 2017. PMID: 29114014 Free PMC article.
-
Functional characterisation of Arabidopsis SPL7 conserved protein domains suggests novel regulatory mechanisms in the Cu deficiency response.BMC Plant Biol. 2014 Aug 30;14:231. doi: 10.1186/s12870-014-0231-5. BMC Plant Biol. 2014. PMID: 25207797 Free PMC article.
-
Toxicity responses of Cu and Cd: the involvement of miRNAs and the transcription factor SPL7.BMC Plant Biol. 2016 Jun 28;16(1):145. doi: 10.1186/s12870-016-0830-4. BMC Plant Biol. 2016. PMID: 27352843 Free PMC article.
-
Transcriptome sequencing identifies SPL7-regulated copper acquisition genes FRO4/FRO5 and the copper dependence of iron homeostasis in Arabidopsis.Plant Cell. 2012 Feb;24(2):738-61. doi: 10.1105/tpc.111.090431. Epub 2012 Feb 28. Plant Cell. 2012. PMID: 22374396 Free PMC article.
-
Copper Trafficking in Plants and Its Implication on Cell Wall Dynamics.Front Plant Sci. 2016 May 6;7:601. doi: 10.3389/fpls.2016.00601. eCollection 2016. Front Plant Sci. 2016. PMID: 27200069 Free PMC article. Review.
Cited by
-
Genome-wide identification and characterization of SPL transcription factor family and their evolution and expression profiling analysis in cotton.Sci Rep. 2018 Jan 15;8(1):762. doi: 10.1038/s41598-017-18673-4. Sci Rep. 2018. PMID: 29335584 Free PMC article.
-
Patterns of genomic divergence in sympatric and allopatric speciation of three Mihoutao (Actinidia) species.Hortic Res. 2022 Mar 3;9:uhac054. doi: 10.1093/hr/uhac054. eCollection 2022. Hortic Res. 2022. PMID: 35591930 Free PMC article.
-
The Arabidopsis KIN17 and its homolog KLP mediate different aspects of plant growth and development.Plant Signal Behav. 2014;9(3):e28634. doi: 10.4161/psb.28634. Epub 2014 Apr 8. Plant Signal Behav. 2014. PMID: 24713636 Free PMC article.
-
Primary nutrient sensors in plants.iScience. 2022 Mar 4;25(4):104029. doi: 10.1016/j.isci.2022.104029. eCollection 2022 Apr 15. iScience. 2022. PMID: 35313690 Free PMC article. Review.
-
CITF1 Functions Downstream of SPL7 to Specifically Regulate Cu Uptake in Arabidopsis.Int J Mol Sci. 2022 Jun 29;23(13):7239. doi: 10.3390/ijms23137239. Int J Mol Sci. 2022. PMID: 35806241 Free PMC article.
References
-
- Agarwal S, Sairam RK, Meena RC, Tyagi A, Srivastava GC. (2006) Effect of excess and deficient levels of iron and copper on oxidative stress and antioxidant enzymes activity in wheat. J Plant Sci 1: 86–97
-
- Angulo JF, Moreau PL, Maunoury R, Laporte J, Hill AM, Bertolotti R, Devoret R. (1989) KIN, a mammalian nuclear protein immunologically related to E. coli RecA protein. Mutat Res 217: 123–134 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

