Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae
- PMID: 24335508
- PMCID: PMC3912081
- DOI: 10.1104/pp.113.230052
Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae
Abstract
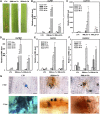
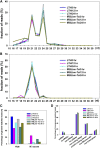
MicroRNAs (miRNAs) are indispensable regulators for development and defense in eukaryotes. However, the miRNA species have not been explored for rice (Oryza sativa) immunity against the blast fungus Magnaporthe oryzae, the most devastating fungal pathogen in rice production worldwide. Here, by deep sequencing small RNA libraries from susceptible and resistant lines in normal conditions and upon M. oryzae infection, we identified a group of known rice miRNAs that were differentially expressed upon M. oryzae infection. They were further classified into three classes based on their expression patterns in the susceptible japonica line Lijiangxin Tuan Hegu and in the resistant line International Rice Blast Line Pyricularia-Kanto51-m-Tsuyuake that contains a single resistance gene locus, Pyricularia-Kanto 51-m (Pikm), within the Lijiangxin Tuan Hegu background. RNA-blot assay of nine of them confirmed sequencing results. Real-time reverse transcription-polymerase chain reaction assay showed that the expression of some target genes was negatively correlated with the expression of miRNAs. Moreover, transgenic rice plants overexpressing miR160a and miR398b displayed enhanced resistance to M. oryzae, as demonstrated by decreased fungal growth, increased hydrogen peroxide accumulation at the infection site, and up-regulated expression of defense-related genes. Taken together, our data indicate that miRNAs are involved in rice immunity against M. oryzae and that overexpression of miR160a or miR398b can enhance rice resistance to the disease.
Figures





Similar articles
-
Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance.PLoS One. 2013;8(3):e59720. doi: 10.1371/journal.pone.0059720. Epub 2013 Mar 27. PLoS One. 2013. PMID: 23544090 Free PMC article.
-
Osa-miR7695 enhances transcriptional priming in defense responses against the rice blast fungus.BMC Plant Biol. 2019 Dec 18;19(1):563. doi: 10.1186/s12870-019-2156-5. BMC Plant Biol. 2019. PMID: 31852430 Free PMC article.
-
The durably resistant rice cultivar Digu activates defence gene expression before the full maturation of Magnaporthe oryzae appressorium.Mol Plant Pathol. 2016 Apr;17(3):354-68. doi: 10.1111/mpp.12286. Epub 2015 Jul 14. Mol Plant Pathol. 2016. PMID: 26095454 Free PMC article.
-
[Recent advances in understanding the innate immune mechanisms and developing new disease resistance breeding strategies against the rice blast fungus Magnaporthe oryzae in rice].Yi Chuan. 2014 Aug;36(8):756-65. doi: 10.3724/SP.J.1005.2014.0756. Yi Chuan. 2014. PMID: 25143273 Review. Chinese.
-
Oscillating Transcriptome during Rice-Magnaporthe Interaction.Curr Issues Mol Biol. 2016;19:99-120. Epub 2015 Sep 11. Curr Issues Mol Biol. 2016. PMID: 26363736 Review.
Cited by
-
miRNA Mediated Regulation and Interaction between Plants and Pathogens.Int J Mol Sci. 2021 Mar 13;22(6):2913. doi: 10.3390/ijms22062913. Int J Mol Sci. 2021. PMID: 33805611 Free PMC article. Review.
-
A natural allele of proteasome maturation factor improves rice resistance to multiple pathogens.Nat Plants. 2023 Feb;9(2):228-237. doi: 10.1038/s41477-022-01327-3. Epub 2023 Jan 16. Nat Plants. 2023. PMID: 36646829
-
Small RNAs Participate in Plant-Virus Interaction and Their Application in Plant Viral Defense.Int J Mol Sci. 2022 Jan 8;23(2):696. doi: 10.3390/ijms23020696. Int J Mol Sci. 2022. PMID: 35054880 Free PMC article. Review.
-
TaMAPK4 Acts as a Positive Regulator in Defense of Wheat Stripe-Rust Infection.Front Plant Sci. 2018 Feb 15;9:152. doi: 10.3389/fpls.2018.00152. eCollection 2018. Front Plant Sci. 2018. PMID: 29527215 Free PMC article.
-
Analysis of miRNAs in Two Wheat Cultivars Infected With Puccinia striiformis f. sp. tritici.Front Plant Sci. 2020 Jan 10;10:1574. doi: 10.3389/fpls.2019.01574. eCollection 2019. Front Plant Sci. 2020. PMID: 31998329 Free PMC article.
References
-
- Alfano JR, Collmer A. (2004) Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol 42: 385–414 - PubMed
-
- Baulcombe D. (2004) RNA silencing in plants. Nature 431: 356–363 - PubMed
-
- Berruyer R, Poussier S, Kankanala P, Mosquera G, Valent B. (2006) Quantitative and qualitative influence of inoculation methods on in planta growth of rice blast fungus. Phytopathology 96: 346–355 - PubMed
-
- Beyer W, Imlay J, Fridovich I. (1991) Superoxide dismutases. Prog Nucleic Acid Res Mol Biol 40: 221–253 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

