A common solution to group 2 influenza virus neutralization
- PMID: 24335589
- PMCID: PMC3890827
- DOI: 10.1073/pnas.1319058110
A common solution to group 2 influenza virus neutralization
Abstract
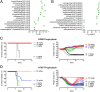
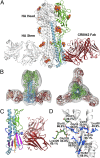
The discovery and characterization of broadly neutralizing antibodies (bnAbs) against influenza viruses have raised hopes for the development of monoclonal antibody (mAb)-based immunotherapy and the design of universal influenza vaccines. Only one human bnAb (CR8020) specifically recognizing group 2 influenza A viruses has been previously characterized that binds to a highly conserved epitope at the base of the hemagglutinin (HA) stem and has neutralizing activity against H3, H7, and H10 viruses. Here, we report a second group 2 bnAb, CR8043, which was derived from a different germ-line gene encoding a highly divergent amino acid sequence. CR8043 has in vitro neutralizing activity against H3 and H10 viruses and protects mice against challenge with a lethal dose of H3N2 and H7N7 viruses. The crystal structure and EM reconstructions of the CR8043-H3 HA complex revealed that CR8043 binds to a site similar to the CR8020 epitope but uses an alternative angle of approach and a distinct set of interactions. The identification of another antibody against the group 2 stem epitope suggests that this conserved site of vulnerability has great potential for design of therapeutics and vaccines.
Keywords: X-ray crystallography; antibody recognition; electron microscopy.
Conflict of interest statement
Conflict of interest statement: R.H.E.F., E.J.M.S., J.J., W.K., M.J., H.J.W.M.K., C.O., E.C.M.B.-v.d.L., T.K., R.V., and J.G. are employees of Crucell Holland B.V.. M.J.K., A.Q.B., T.B., and H.S. are employees of AIMM Therapeutics.
Figures



References
-
- World Health Organization (WHO) Fact Sheet 211: Influenza. Geneva: World Health Organization; 2009.
-
- Potter CW. A history of influenza. J Appl Microbiol. 2001;91(4):572–579. - PubMed
-
- Stephenson I, et al. Neuraminidase inhibitor resistance after oseltamivir treatment of acute influenza A and B in children. Clin Infect Dis. 2009;48(4):389–396. - PubMed
-
- Carrat F, Flahault A. Influenza vaccine: The challenge of antigenic drift. Vaccine. 2007;25(39–40):6852–6862. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

