Fluorescent probes for nucleic Acid visualization in fixed and live cells
- PMID: 24335616
- PMCID: PMC6270009
- DOI: 10.3390/molecules181215357
Fluorescent probes for nucleic Acid visualization in fixed and live cells
Abstract
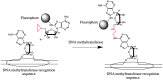
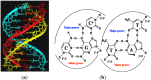
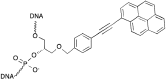
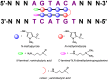
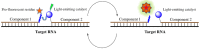
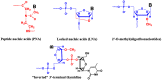
This review analyses the literature concerning non-fluorescent and fluorescent probes for nucleic acid imaging in fixed and living cells from the point of view of their suitability for imaging intracellular native RNA and DNA. Attention is mainly paid to fluorescent probes for fluorescence microscopy imaging. Requirements for the target-binding part and the fluorophore making up the probe are formulated. In the case of native double-stranded DNA, structure-specific and sequence-specific probes are discussed. Among the latest, three classes of dsDNA-targeting molecules are described: (i) sequence-specific peptides and proteins; (ii) triplex-forming oligonucleotides and (iii) polyamide oligo(N-methylpyrrole/N-methylimidazole) minor groove binders. Polyamides seem to be the most promising targeting agents for fluorescent probe design, however, some technical problems remain to be solved, such as the relatively low sequence specificity and the high background fluorescence inside the cells. Several examples of fluorescent probe applications for DNA imaging in fixed and living cells are cited. In the case of intracellular RNA, only modified oligonucleotides can provide such sequence-specific imaging. Several approaches for designing fluorescent probes are considered: linear fluorescent probes based on modified oligonucleotide analogs, molecular beacons, binary fluorescent probes and template-directed reactions with fluorescence probe formation, FRET donor-acceptor pairs, pyrene excimers, aptamers and others. The suitability of all these methods for living cell applications is discussed.
Figures

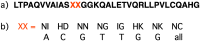
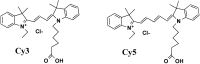
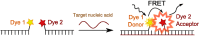

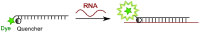
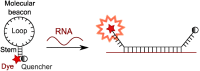
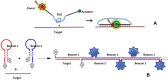

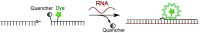
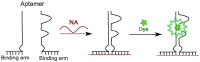
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

